3OMX
 
 | | Crystal structure of Ssu72 with vanadate complex | | 分子名称: | CG14216, VANADATE ION | | 著者 | Zhang, Y, Zhang, M, Zhang, Y. | | 登録日 | 2010-08-27 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3366 Å) | | 主引用文献 | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
5D4D
 
 | |
5TIG
 
 | |
5D4C
 
 | |
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | 登録日 | 2004-05-28 | | 公開日 | 2004-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1VZU
 
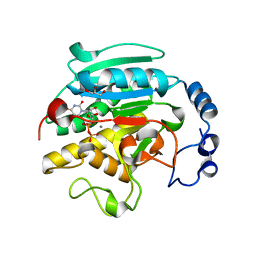 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | 登録日 | 2004-05-27 | | 公開日 | 2004-07-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
7EN5
 
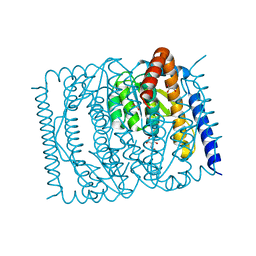 | | The crystal structure of Escherichia coli MurR in complex with N-acetylglucosamine-6-phosphate | | 分子名称: | 2-METHOXYETHANOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhang, Y, Chen, W, Ji, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7EN7
 
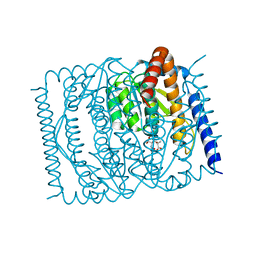 | | The crystal structure of Escherichia coli MurR in complex with N-acetylmuramic-acid-6-phosphate | | 分子名称: | (2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2,5-bis(oxidanyl)-6-(phosphonooxymethyl)oxan-4-yl]oxypropanoic acid, HTH-type transcriptional regulator MurR | | 著者 | Zhang, Y, Chen, W, Ji, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Molecular basis for cell-wall recycling regulation by transcriptional repressor MurR in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7KHJ
 
 | |
7KHK
 
 | |
7KHG
 
 | | Crystal structure of KIT kinase domain with a small molecule inhibitor, PLX3397 | | 分子名称: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Mast/stem cell growth factor receptor Kit | | 著者 | Zhang, Y. | | 登録日 | 2020-10-21 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Association of Combination of Conformation-Specific KIT Inhibitors With Clinical Benefit in Patients With Refractory Gastrointestinal Stromal Tumors: A Phase 1b/2a Nonrandomized Clinical Trial.
Jama Oncol, 7, 2021
|
|
5KXF
 
 | |
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
9ATN
 
 | |
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-12 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
5JKF
 
 | | Crystal structure of esterase E22 | | 分子名称: | Esterase E22 | | 著者 | Zhang, Y, Wang, P, Yao, Q. | | 登録日 | 2016-04-26 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.393 Å) | | 主引用文献 | Structural basis for substrate recognition and catalysis of a novel esterase E22 with a homoserine transacetylase-like fold
To Be Published
|
|
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | 分子名称: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
5JKJ
 
 | |
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-22 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1PQN
 
 | | dominant negative human hDim1 (hDim1 1-128) | | 分子名称: | Spliceosomal U5 snRNP-specific 15 kDa protein | | 著者 | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | 登録日 | 2003-06-18 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
7CZH
 
 | | PL24 ulvan lyase-Uly1 | | 分子名称: | CALCIUM ION, GLYCEROL, Uly1 | | 著者 | Zhang, Y.Z, Chen, X.L, Dong, F, Xu, F. | | 登録日 | 2020-09-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Mechanistic Insights into Substrate Recognition and Catalysis of a New Ulvan Lyase of Polysaccharide Lyase Family 24.
Appl.Environ.Microbiol., 87, 2021
|
|
4LA2
 
 | |
4LA3
 
 | |
1TGE
 
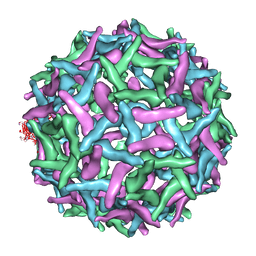 | | The structure of immature Dengue virus at 12.5 angstrom | | 分子名称: | envelope glycoprotein | | 著者 | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | 登録日 | 2004-05-28 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
