4C6T
 
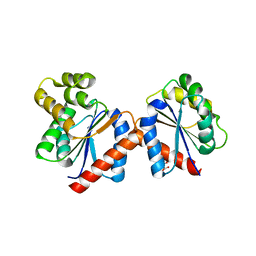 | | Crystal structure of the RPS4 and RRS1 TIR domain heterodimer | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4, MALONIC ACID, PROBABLE WRKY TRANSCRIPTION FACTOR 52 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2DI4
 
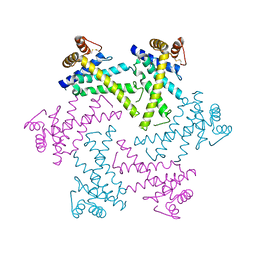 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2BJV
 
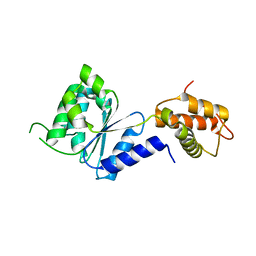 | | Crystal Structure of PspF(1-275) R168A mutant | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2C98
 
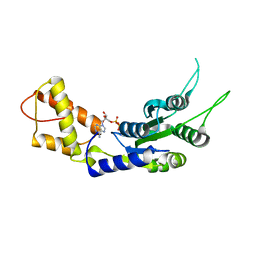 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C99
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
4R2E
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
2C96
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
4R2R
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2P
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2A
 
 | | Egr1/Zif268 zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
2C9C
 
 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
4R2D
 
 | | Egr1/Zif268 zinc fingers in complex with formylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2C
 
 | | Egr1/Zif268 zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
4R2S
 
 | | Wilms Tumor Protein (WT1) Q369P zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
6NT6
 
 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
4R2Q
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with formylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
2BJW
 
 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
6NT7
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound dimeric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | Descriptor: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
6NT8
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6VXK
 
 | | Cryo-EM Structure of the full-length A39R/PlexinC1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, Semaphorin-like protein 139 | | Authors: | Kuo, Y.-C, Chen, H, Shang, G, Uchikawa, E, Tian, H, Bai, X, Zhang, X. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Nat Commun, 11, 2020
|
|
7M0R
 
 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
