5WTA
 
 | | Crystal Structure of Staphylococcus aureus SdrE apo form | | 分子名称: | Serine-aspartate repeat-containing protein E | | 著者 | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | 登録日 | 2016-12-10 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
6IFU
 
 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | 分子名称: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | 分子名称: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2019-01-23 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6M3W
 
 | |
3T9N
 
 | | Crystal structure of a membrane protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | 著者 | Yang, M, Zhang, X, Ge, J, Wang, J. | | 登録日 | 2011-08-03 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7EGK
 
 | | Bicarbonate transporter complex SbtA-SbtB bound to AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein SbtB, SODIUM ION, ... | | 著者 | Fang, S, Huang, X, Zhang, X, Zhang, P. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EGL
 
 | | Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | | 分子名称: | BICARBONATE ION, Membrane-associated protein SbtB, SODIUM ION, ... | | 著者 | Fang, S, Huang, X, Zhang, X, Zhang, P. | | 登録日 | 2021-03-24 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WTB
 
 | | Complex Structure of Staphylococcus aureus SdrE with human complement factor H | | 分子名称: | Peptide from Complement factor H, Serine-aspartate repeat-containing protein E | | 著者 | Wu, M, Zhang, Y, Hang, T, Wang, C, Yang, Y, Zang, J, Zhang, M, Zhang, X. | | 登録日 | 2016-12-10 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Staphylococcus aureus SdrE captures complement factor H's C-terminus via a novel 'close, dock, lock and latch' mechanism for complement evasion
Biochem. J., 474, 2017
|
|
7LCI
 
 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | 分子名称: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | 登録日 | 2021-01-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
8WQL
 
 | | In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | You, X, Zhang, X, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2023-10-11 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of in situ PBS-PSII supercomplex at 3.5 Angstroms resolution.
To Be Published
|
|
8IAB
 
 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | 著者 | Yang, Z, Zhang, X, Zhang, P. | | 登録日 | 2023-02-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | 著者 | Yang, Z, Zhang, X, Zhang, P. | | 登録日 | 2023-02-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
4MGG
 
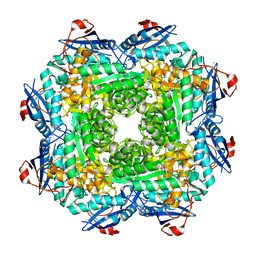 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | 著者 | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-08-28 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
4M8N
 
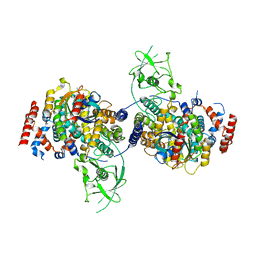 | | Crystal Structure of PlexinC1/Rap1B Complex | | 分子名称: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | 登録日 | 2013-08-13 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.294 Å) | | 主引用文献 | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
4HIN
 
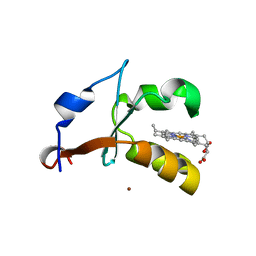 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | 分子名称: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | 登録日 | 2012-10-11 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
5YF0
 
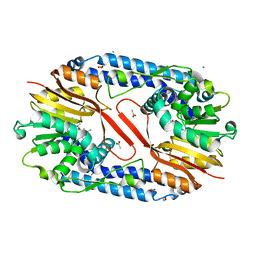 | | Crystal structure of CARNMT1 bound to SAM | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Cao, R, Zhang, X, Li, H. | | 登録日 | 2017-09-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular basis for histidine N1 position-specific methylation by CARNMT1.
Cell Res., 28, 2018
|
|
4IP3
 
 | | Complex structure of OspI and Ubc13 | | 分子名称: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | 著者 | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | 登録日 | 2013-01-09 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | 著者 | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | 登録日 | 2013-06-25 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7YE6
 
 | | BAM-EspP complex structure with BamA-N427C/EspP-R1297C mutations in nanodisc | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
7YE4
 
 | | BAM-EspP complex structure with BamA-G431C and G781C/EspP-N1293C and A1043C mutations in nanodisc | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Shen, C, Chang, S, Luo, Q, Zhang, Z, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhang, X, Tang, X, Dong, H. | | 登録日 | 2022-07-05 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
4M8M
 
 | |
