7DYB
 
 | | Thermotoga maritima ferritin mutant-FLAL-L | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7BEG
 
 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
7YL3
 
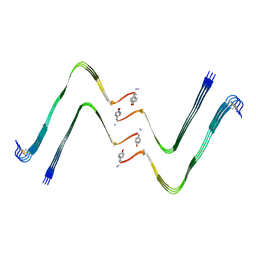 | | Structure of hIAPP-TF-type1 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X, Wang, Y, Zhu, P. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YL0
 
 | | Structure of hIAPP-TF-type2 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X, Wang, Y, Zhu, P. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
6LS0
 
 | |
7YL7
 
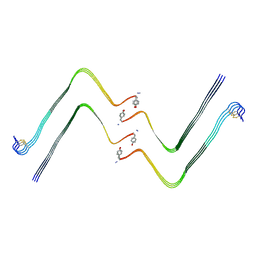 | | Structure of hIAPP-TF-type3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YKW
 
 | |
4PX7
 
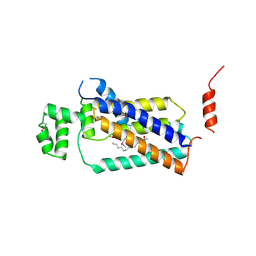 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PEJ
 
 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
7DAW
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | Descriptor: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
4M8M
 
 | |
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QD7
 
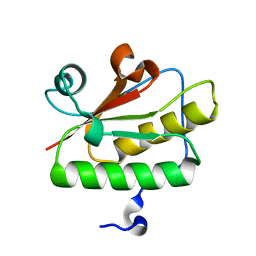 | | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli | | Descriptor: | Uncharacterized protein ydaL | | Authors: | Gui, W.J, Qu, Q.H, Chen, Y.Y, Wang, M, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2011-01-18 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli.
J.Struct.Biol., 174, 2011
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
1F3Y
 
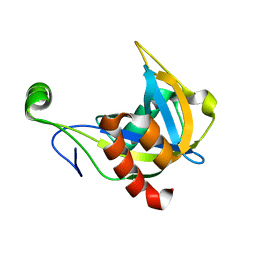 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | Descriptor: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | Authors: | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
4EJY
 
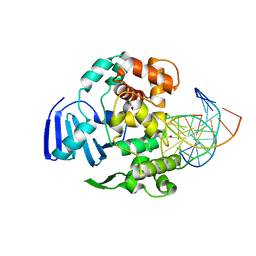 | | Structure of MBOgg1 in complex with high affinity DNA ligand | | Descriptor: | 3-Methyladenine DNA glycosylase, DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), ... | | Authors: | Jiang, T, Yu, H.J, Bi, L.J, Zhang, X.E, Yang, M.Z. | | Deposit date: | 2012-04-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of MBOgg1 in complex with two abasic DNA ligands
J.Struct.Biol., 181, 2013
|
|
7CCR
 
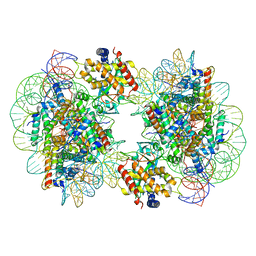 | | Structure of the 2:2 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
 | | Structure of the 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
4EJZ
 
 | | Structure of MBOgg1 in complex with low affinity DNA ligand | | Descriptor: | 3-Methyladenine DNA glycosylase, DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*T*GP*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3') | | Authors: | Jiang, T, Yu, H.J, Bi, L.J, Zhang, X.E, Yang, M.Z. | | Deposit date: | 2012-04-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of MBOgg1 in complex with two abasic DNA ligands
J.Struct.Biol., 181, 2013
|
|
6KD3
 
 | |
