3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-10-23 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
4E0R
 
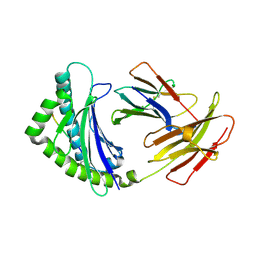 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
7YFM
 
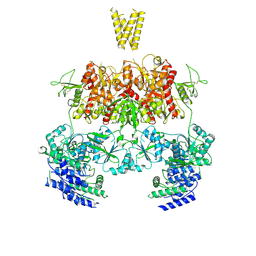 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
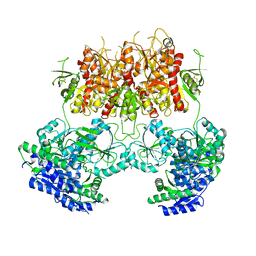 | |
5EGF
 
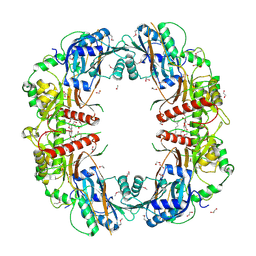 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
7SBQ
 
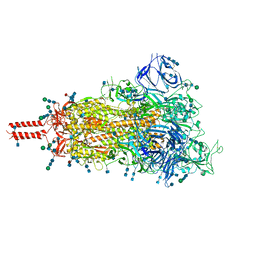 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBP
 
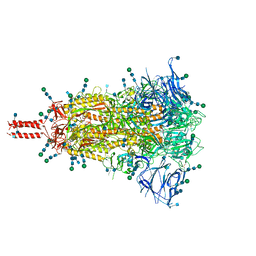 | | Closed state of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBR
 
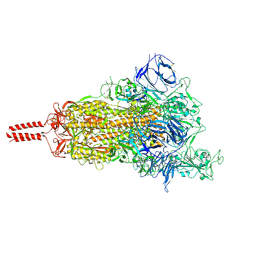 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBO
 
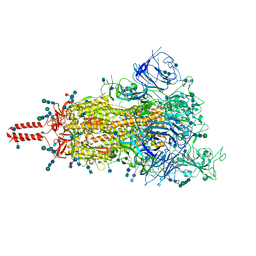 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBT
 
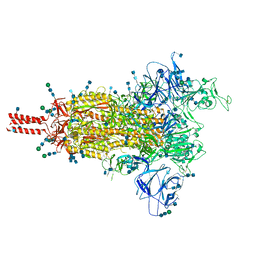 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBK
 
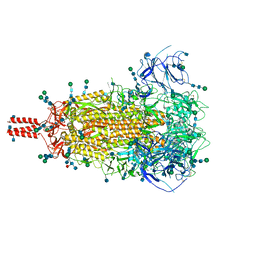 | | Closed state of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBS
 
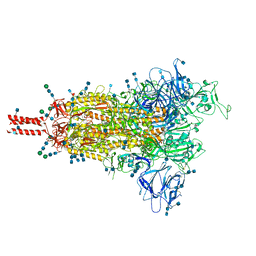 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
7SBL
 
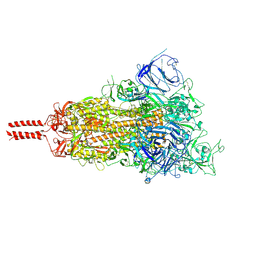 | | One RBD-up 1 of pre-fusion SARS-CoV-2 Delta variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
2RBE
 
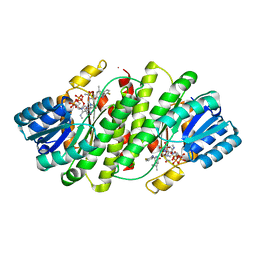 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1Y5M
 
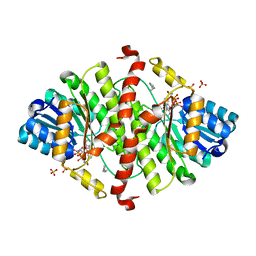 | | The crystal structure of murine 11b-hydroxysteroid dehydrogenase: an important therapeutic target for diabetes | | Descriptor: | Corticosteroid 11-beta-dehydrogenase, isozyme 1, N-OCTANE, ... | | Authors: | Zhang, J, Osslund, T.D, Plant, M.H, Clogston, C.L, Nybo, R.E, Xiong, F, Delaney, J.M, Jordan, S.R. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Murine 11-Hydroxysteroid Dehydrogenase 1: An Important Therapeutic Target for Diabetes
Biochemistry, 44, 2005
|
|
3EY4
 
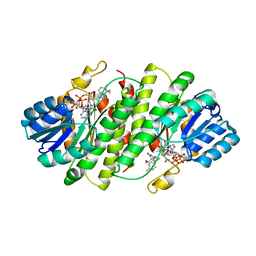 | | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model | | Descriptor: | (5S)-2-{[(1S)-1-(4-fluorophenyl)ethyl]amino}-5-(1-hydroxy-1-methylethyl)-5-methyl-1,3-thiazol-4(5H)-one, 11-beta-Hydroxysteroid Dehydrogenase 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J.D, Jordan, S.R, Li, V. | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model
To be Published, 2008
|
|
4L2Y
 
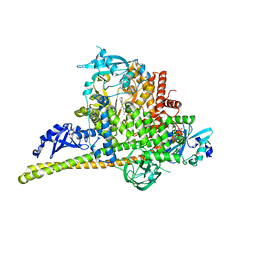 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L1B
 
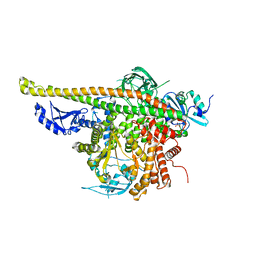 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
7YUK
 
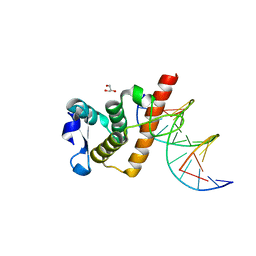 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
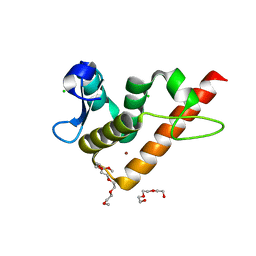 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
6XR8
 
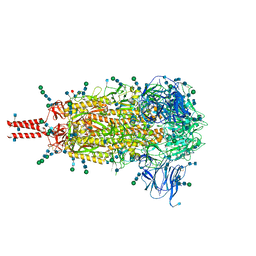 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
5Z9Y
 
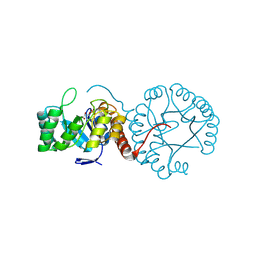 | | Crystal structure of Mycobacterium tuberculosis thiazole synthase (ThiG) complexed with DXP | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Thiazole synthase | | Authors: | Zhang, J, Zhang, B, Zhao, Y, Yang, X, Huang, M, Cui, P, Zhang, W, Li, J, Zhang, Y. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Snapshots of catalysis: Structure of covalently bound substrate trapped in Mycobacterium tuberculosis thiazole synthase (ThiG).
Biochem. Biophys. Res. Commun., 497, 2018
|
|
6XRA
 
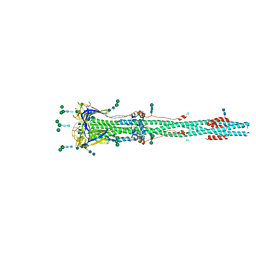 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
4L23
 
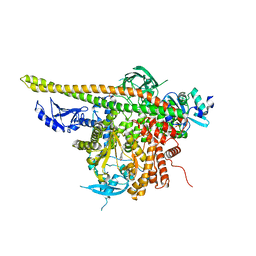 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
3J02
 
 | | Lidless D386A Mm-cpn in the pre-hydrolysis ATP-bound state | | Descriptor: | Lidless D386A Mm-cpn variant | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
