2WTK
 
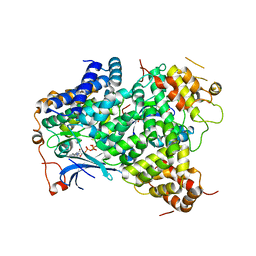 | |
3GNI
 
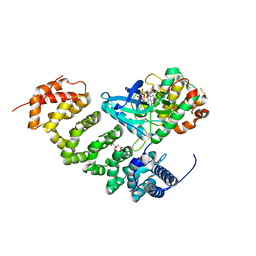 | | Structure of STRAD and MO25 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Protein Mo25, ... | | Authors: | Zeqiraj, E, Goldie, S, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2009-03-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor
Plos Biol., 7, 2009
|
|
5CW6
 
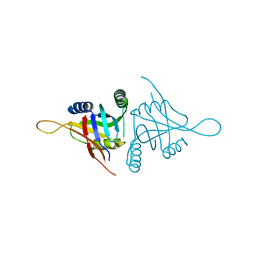 | | Structure of metal dependent enzyme DrBRCC36 | | Descriptor: | DrBRCC36, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.193 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW3
 
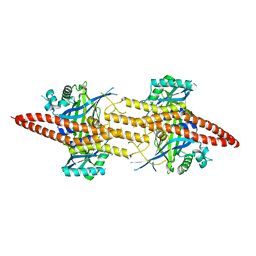 | | Structure of CfBRCC36-CfKIAA0157 complex (Zn Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW4
 
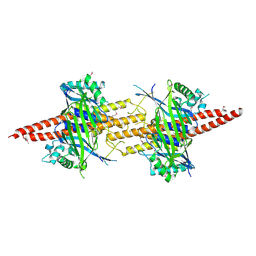 | | Structure of CfBRCC36-CfKIAA0157 complex (Selenium Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, GLYCEROL, Protein FAM175B, ... | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW5
 
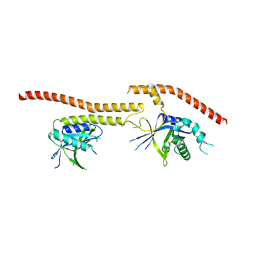 | | Structure of CfBRCC36-CfKIAA0157 complex (QSQ mutant) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.736 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | Descriptor: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | Authors: | Zeqiraj, E, Judd, A, Sicheri, F. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZBN
 
 | |
8QFD
 
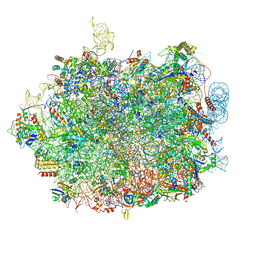 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Makhlouf, L, Kulathu, Y, Zeqiraj, E. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8OFF
 
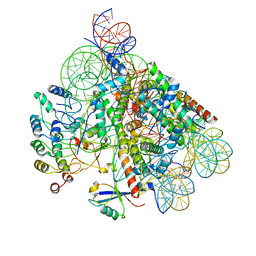 | | Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map1) | | Descriptor: | BRCA1 associated RING domain 1, DNA (142-MER), Histone H2A type 1, ... | | Authors: | Foglizzo, M, Burdett, H, Wilson, M.D, Zeqiraj, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BRCA1-BARD1 combines multiple chromatin recognition modules to bridge nascent nucleosomes.
Nucleic Acids Res., 51, 2023
|
|
8RHN
 
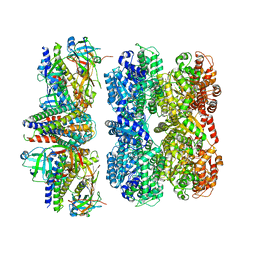 | | Structure of the 55LCC ATPase complex | | Descriptor: | ATPase family gene 2 protein homolog A, ATPase family gene 2 protein homolog B, Cyclin-dependent kinase 2-interacting protein, ... | | Authors: | Foglizzo, M, Degtjarik, O, Zeqiraj, E. | | Deposit date: | 2023-12-15 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
8QFC
 
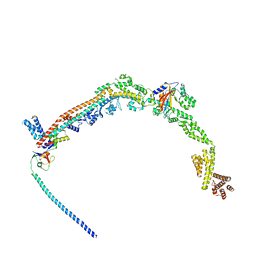 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | Authors: | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8CIH
 
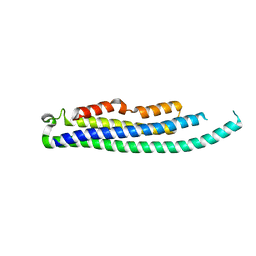 | | Structure of FL CINP | | Descriptor: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | Authors: | Foglizzo, M, Zeqiraj, E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
6R8F
 
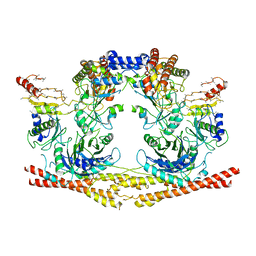 | | Cryo-EM structure of the Human BRISC-SHMT2 complex | | Descriptor: | BRISC and BRCA1-A complex member 2,BRCC45 (BRE, BRISC and BRCA1-A complex member 2), BRISC complex subunit Abraxas 2, ... | | Authors: | Walden, M, Hesketh, E, Tian, L, Ranson, N.A, Greenberg, R.A, Zeqiraj, E. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Metabolic control of BRISC-SHMT2 assembly regulates immune signalling.
Nature, 570, 2019
|
|
3TZM
 
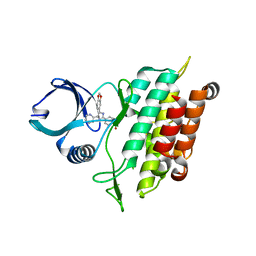 | | TGF-beta Receptor type 1 in complex with SB431542 | | Descriptor: | 4-[5-(1,3-benzodioxol-5-yl)-4-(pyridin-2-yl)-1H-imidazol-2-yl]benzamide, TGF-beta receptor type-1 | | Authors: | Ogunjimi, A.A, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Specificity of TGFbeta Family Receptor Small Molecule Inhibitors
Cell Signal, 24, 2012
|
|
4O1O
 
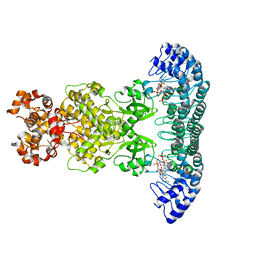 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4NKH
 
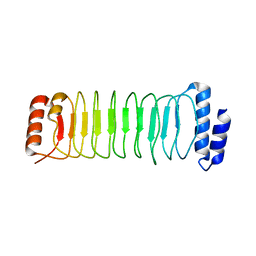 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4NKG
 
 | | Crystal structure of SspH1 LRR domain in complex PKN1 HR1b domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1, HEXANE-1,6-DIOL, Serine/threonine-protein kinase N1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
4O1P
 
 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
