2MVZ
 
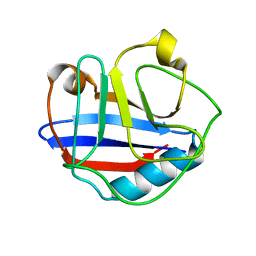 | | Solution Structure for Cyclophilin A from Geobacillus Kaustophilus | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Holliday, M.J, Isern, N.G, Geoffrey, A.S, Zhang, F, Eisenmesser, E.Z. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of GeoCyp: A Thermophilic Cyclophilin with a Novel Substrate Binding Mechanism That Functions Efficiently at Low Temperatures.
Biochemistry, 54, 2015
|
|
8FC7
 
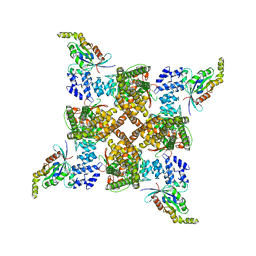 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK2798745 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8UYO
 
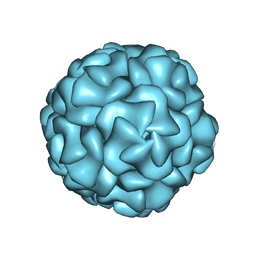 | |
3E63
 
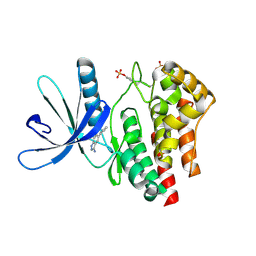 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 5-phenyl-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E62
 
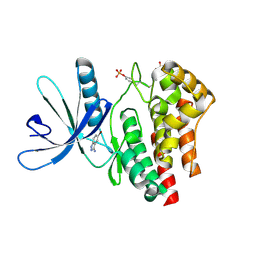 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 5-bromo-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E64
 
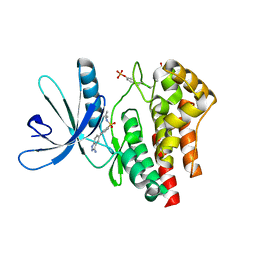 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 4-(3-amino-1H-indazol-5-yl)-N-tert-butylbenzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2LLF
 
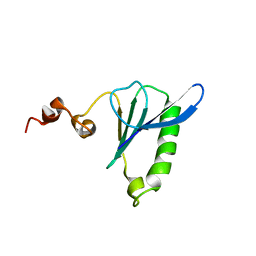 | | Sixth Gelsolin-like domain of villin in 5 mM CaCl2 | | Descriptor: | Villin-1 | | Authors: | Pfaff, D.A, Brockerman, J, Fedechkin, S, Burns, L, Zhang, F, Mcknight, C, Smirnov, S.L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Gelsolin-like activation of villin: calcium sensitivity of the long helix in domain 6.
Biochemistry, 52, 2013
|
|
5GYL
 
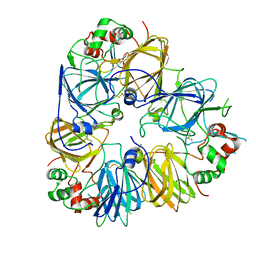 | | Structure of Cicer arietinum 11S gloubulin | | Descriptor: | legumin-like protein | | Authors: | Zhou, A, Zhang, F. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization and crystallographic studies of a novel chickpea 11S globulin.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3DIP
 
 | | Crystal structure of an enolase protein from the environmental genome shotgun sequencing of the Sargasso Sea | | Descriptor: | SULFATE ION, enolase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an enolase protein from the environmental genome shotgun sequencing of the Sargasso Sea
To be Published
|
|
8W1P
 
 | |
2P2U
 
 | | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris | | Descriptor: | Host-nuclease inhibitor protein Gam, putative | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Zhang, F, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of putative host-nuclease inhibitor protein Gam from Desulfovibrio vulgaris
To be Published
|
|
2P1G
 
 | | Crystal structure of a putative xylanase from Bacteroides fragilis | | Descriptor: | Putative xylanase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative xylanase from Bacteroides fragilis
To be Published
|
|
5UF6
 
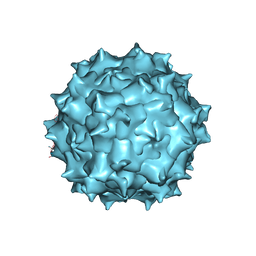 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
5UMO
 
 | |
8GH6
 
 | |
8GKH
 
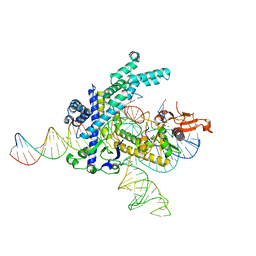 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2023-03-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
6RPJ
 
 | |
7E3X
 
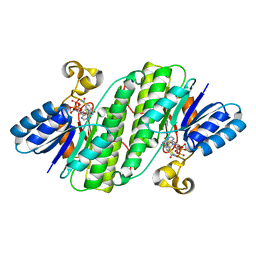 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
7VT1
 
 | |
