1LR7
 
 | |
6Q38
 
 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
5FOV
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTG TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, FHTG PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FPK
 
 | | MONOMERIC RADA IN COMPLEX WITH FATA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTG PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOX
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHAA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHAA PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
5FOW
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
1LR9
 
 | |
1LR8
 
 | |
5BXO
 
 | | Human Tankyrase-2 in Complex with Macrocyclised Extended Peptide cp4n2m3 | | Descriptor: | 4,4'-propane-1,3-diylbis(1-methyl-1H-1,2,3-triazole), DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Xu, W, Fischer, G, Hyvonen, M, Itzhaki, L. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-29 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Macrocyclized Extended Peptides: Inhibiting the Substrate-Recognition Domain of Tankyrase.
J.Am.Chem.Soc., 139, 2017
|
|
6YPN
 
 | |
6YJE
 
 | | Plasmoodium vivax phosphoglycerate kinase bound to nitrofuran inhibitor from PEG3350 and ammonium acetate at pH 5.5 | | Descriptor: | (2~{S})-2-(5-nitrofuran-2-yl)-2,3,5,6,7,8-hexahydro-1~{H}-[1]benzothiolo[2,3-d]pyrimidin-4-one, Phosphoglycerate kinase | | Authors: | Blaszczyk, B.K, Hyvonen, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Phosphoglycerate Kinase as a potential target for antimalarial therapy
to be published
|
|
7FS3
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 15 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-4,4',5-trihydroxy[1,1'-biphenyl]-2-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSA
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 24 | | Descriptor: | (1M)-6-[4-(3-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl][1,1'-biphenyl]-3,3',4,4'-tetrol, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRV
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 3 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4,4'-sulfonyldi(benzene-1,2-diol), MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS1
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 11 | | Descriptor: | (1P)-N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-3',4,4',5-tetrahydroxy[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS0
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 8 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3,4-dihydroxy-N-{2-[4-(3-hydroxybenzene-1-sulfonyl)phenyl]ethyl}benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRY
 
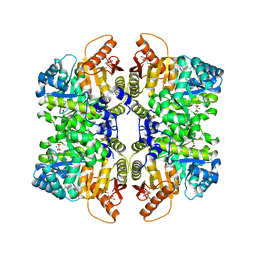 | | Structure of liver pyruvate kinase in complex with allosteric modulator 6 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{2-[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]ethyl}-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS5
 
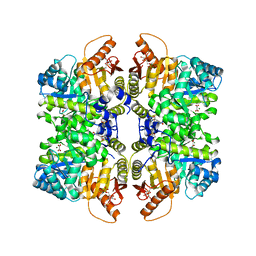 | | Structure of liver pyruvate kinase in complex with allosteric modulator 17 | | Descriptor: | (10aP)-6-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-2,3,8,9-tetrahydroxy-5lambda~6~-dibenzo[c,e][1,2]thiazine-5,5(6H)-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS7
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 20 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-N-(3,4-dihydroxyphenyl)-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS6
 
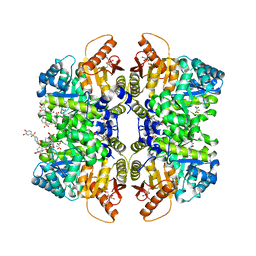 | | Structure of liver pyruvate kinase in complex with allosteric modulator 18 | | Descriptor: | (10aM)-6-{2-[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]ethyl}-2,3,8,9-tetrahydroxy-5lambda~6~-dibenzo[c,e][1,2]thiazine-5,5(6H)-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
5DPV
 
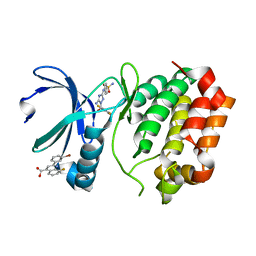 | | Aurora A Kinase in Complex with AA35 and JNJ-7706621 in Space Group P6122 | | Descriptor: | 2-(3-bromophenyl)-8-fluoroquinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | Authors: | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
7FRZ
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 7 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3,4-dihydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSB
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 41 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRX
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRW
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 4 | | Descriptor: | (10aM)-3,8,9-trihydroxy-6H-6lambda~6~-dibenzo[c,e][1,2]oxathiine-6,6-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
