5T5G
 
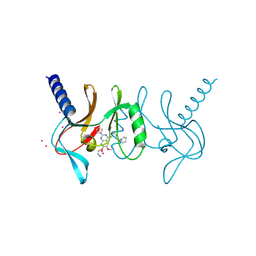 | | human SETD8 in complex with MS2177 | | Descriptor: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
2K4K
 
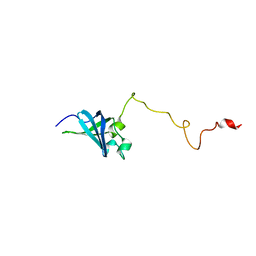 | | Solution structure of GSP13 from Bacillus subtilis | | Descriptor: | General stress protein 13 | | Authors: | Yu, W, Yu, B, Hu, J, Jin, C, Xia, B. | | Deposit date: | 2008-06-13 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins.
J.Biomol.Nmr, 43, 2009
|
|
7Q1K
 
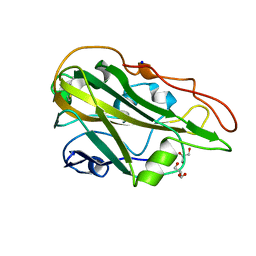 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | Deposit date: | 2021-10-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
5JUW
 
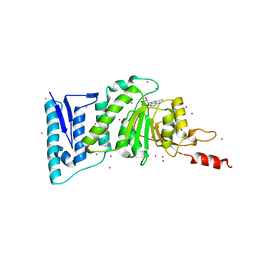 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
5TH7
 
 | | Complex of SETD8 with MS453 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[6,7-dimethoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)propanamide, N-lysine methyltransferase KMT5A, ... | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
4IJ8
 
 | | Crystal structure of the complex of SETD8 with SAM | | Descriptor: | N-lysine methyltransferase SETD8, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Yu, W, Tempel, W, Li, Y, El Bakkouri, M, Shapira, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of SETD8 with SAM
To be Published
|
|
3SX0
 
 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
2HF6
 
 | | Solution structure of human zeta-COP | | Descriptor: | Coatomer subunit zeta-1 | | Authors: | Yu, W, Jin, C, Xia, B. | | Deposit date: | 2006-06-23 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats
J.Mol.Biol., 386, 2009
|
|
3UWP
 
 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4IPN
 
 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
7V1T
 
 | | A dual Inhibitor Against Main Protease | | Descriptor: | 3C-like proteinase, 5,8-bis(oxidanylidene)-7-[(2-piperazin-1-ylphenyl)amino]naphthalene-1-sulfonamide | | Authors: | Yu, W.Y, Xiao, Y.B, Zhao, Y.C. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | SARS-CoV-2 Mpro complex with inhibitor
To Be Published
|
|
7MOY
 
 | | Structure of HDAC2 in complex with an inhibitor (compound 19) | | Descriptor: | (1S)-6-ethyl-N-{(1S)-1-[5-(2-ethyl-1-oxo-1,2-dihydroisoquinolin-6-yl)-1H-imidazol-2-yl]-7,7-dihydroxynonyl}-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOT
 
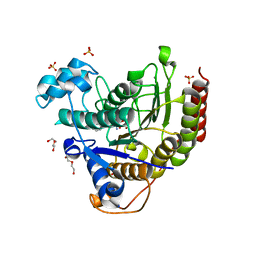 | | Structure of HDAC2 in complex with an inhibitor (compound 9) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOZ
 
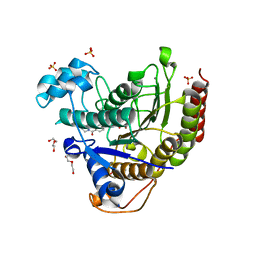 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 25) | | Descriptor: | (1R,3S,6S,18R,27R)-6-(6,6-dihydroxyoctyl)-5,8,18,27,34-pentaazahexacyclo[25.2.2.1~7,10~.1~11,15~.1~14,18~.0~1,3~]tetratriaconta-7,9,11(33),12,14,16-hexaene-4,32-dione (non-preferred name), CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOS
 
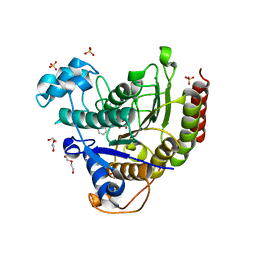 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOX
 
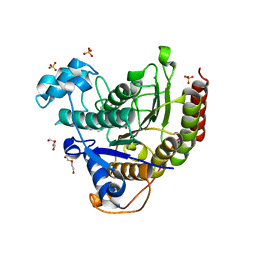 | | Structure of HDAC2 in complex with an inhibitor (compound 14) | | Descriptor: | (1S)-N-[(1S)-7,7-dihydroxy-1-{4-[(1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-6-yl]-1H-imidazol-2-yl}nonyl]-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
8XVF
 
 | |
6WBW
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WBZ
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7OWG
 
 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
5MTQ
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT511 | | Descriptor: | 2-[4-[(4-cyclohexyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5MTP
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT514 | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5MTR
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT512 | | Descriptor: | 2-[4-[(4-cyclopentyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
1ZB7
 
 | | Crystal Structure of Botulinum Neurotoxin Type G Light Chain | | Descriptor: | CITRATE ANION, ZINC ION, neurotoxin | | Authors: | Arndt, J.W, Yu, W, Bi, F, Stevens, R.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of botulinum neurotoxin type g light chain: serotype divergence in substrate recognition
Biochemistry, 44, 2005
|
|
