8GR2
 
 | |
6M58
 
 | |
5ZWR
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, Est-Y29, GLYCEROL | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWQ
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29, GLYCEROL, ethyl (2S)-2-[3-(benzenecarbonyl)phenyl]propanoate | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
4XPL
 
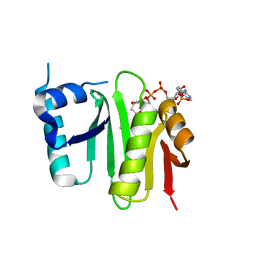 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
4XPK
 
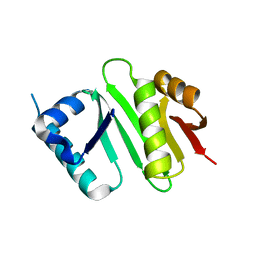 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH | | Descriptor: | N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
8ZAH
 
 | |
8ZAK
 
 | |
7BYK
 
 | |
8XJG
 
 | |
8XJE
 
 | |
6KVX
 
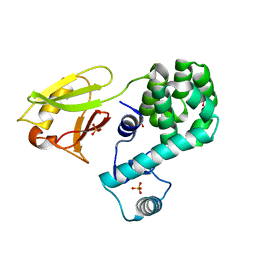 | | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Endolysin,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Park, J, Lee, G, Yoon, S, Min, C.K, Kim, T.G, Yamamoto, T, Kim, D.H, Lee, K.W, Eom, S.H. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the N-terminal domain single mutant (D119A) of the human mitochondrial calcium uniporter fused with T4 lysozyme
Sci Rep, 2020
|
|
7C7Z
 
 | |
8YJX
 
 | |
8K3F
 
 | |
2VR5
 
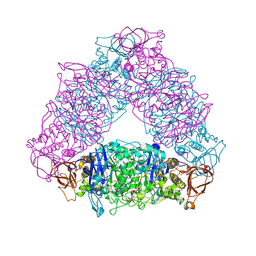 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
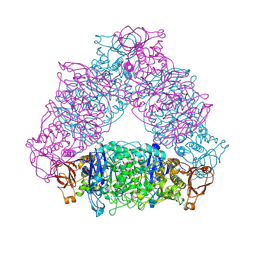 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
7C51
 
 | |
8IEU
 
 | |
8IEV
 
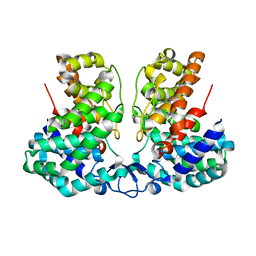 | |
8J56
 
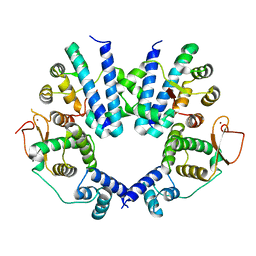 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
5JE8
 
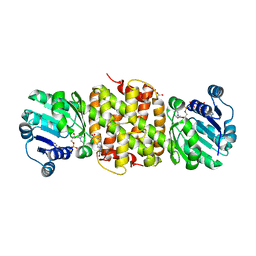 | |
5GLF
 
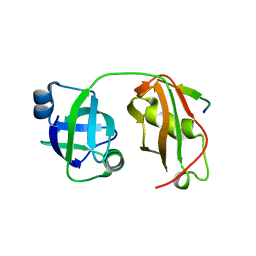 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
5H5V
 
 | |
7C50
 
 | |
