1Q1Y
 
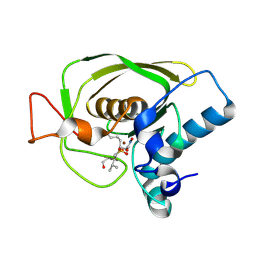 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
3DUV
 
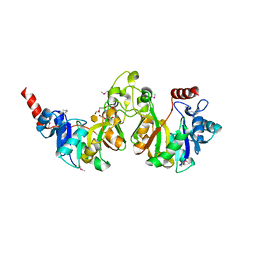 | | Crystal structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Haemophilus influenzae complexed with the substrate 3-deoxy-manno-octulosonate in the-configuration | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL | | Authors: | Yoon, H.J, Ku, M.J, Mikami, B, Suh, S.W. | | Deposit date: | 2008-07-18 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Haemophilus influenzae complexed with the substrate 3-deoxy-manno-octulosonate in the beta-configuration.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3ND5
 
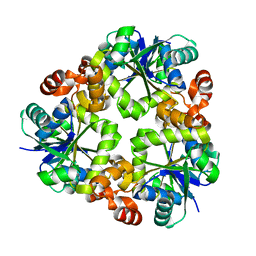 | |
3ND6
 
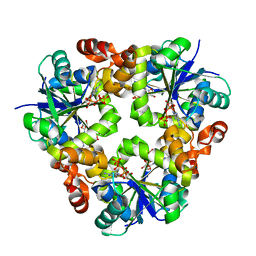 | |
3ND7
 
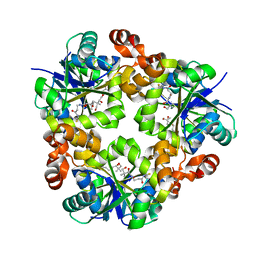 | | Crystal structure of phosphopantetheine adenylyltransferase from Enterococcus faecalis in the ligand-unbound state and in complex with ATP and pantetheine | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, Phosphopantetheine adenylyltransferase | | Authors: | Yoon, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2010-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphopantetheine adenylyltransferase from Enterococcus faecalis in the ligand-unbound state and in complex with ATP and pantetheine
Mol.Cells, 32, 2011
|
|
1CQY
 
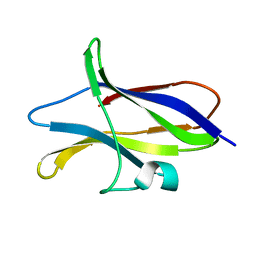 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
2RL2
 
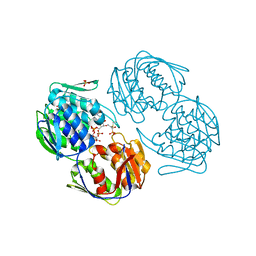 | |
2RL1
 
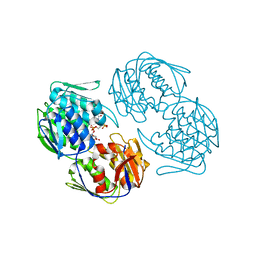 | |
1YUL
 
 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, Probable nicotinate-nucleotide adenylyltransferase | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
1YUN
 
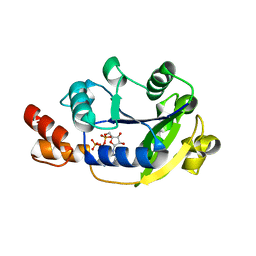 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
1YUM
 
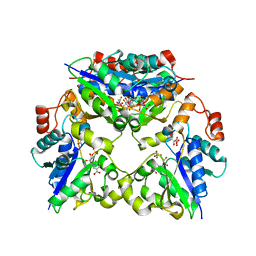 | | Crystal Structure of Nicotinic Acid Mononucleotide Adenylyltransferase from Pseudomonas aeruginosa | | Descriptor: | 'Probable nicotinate-nucleotide adenylyltransferase, CITRIC ACID, NICOTINATE MONONUCLEOTIDE | | Authors: | Yoon, H.J, Kim, H.L, Mikami, B, Suh, S.W. | | Deposit date: | 2005-02-14 | | Release date: | 2005-11-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nicotinic acid mononucleotide adenylyltransferase from Pseudomonas aeruginosa in its Apo and substrate-complexed forms reveals a fully open conformation
J.Mol.Biol., 351, 2005
|
|
3SXN
 
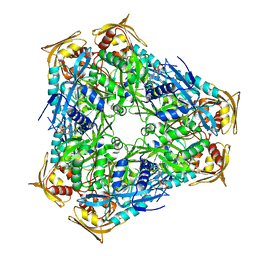 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4RNZ
 
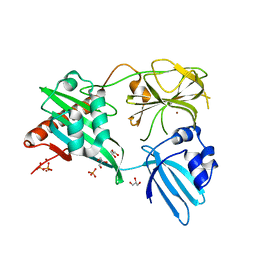 | | Structure of Helicobacter pylori Csd3 from the hexagonal crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3GIO
 
 | | Crystal structure of the TNF-alpha inducing protein (Tip alpha) from Helicobacter pylori | | Descriptor: | Putative uncharacterized protein | | Authors: | Jang, J.Y, Yoon, H.J, Yoon, J.Y, Kim, H.S, Lee, S.J, Kim, K.H, Kim, D.J, Han, B.G, Lee, B.I, Jang, S, Suh, S.W. | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the TNF-alpha-Inducing Protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-Binding Activity.
J.Mol.Biol., 2009
|
|
4Y4V
 
 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3F3M
 
 | |
1PX8
 
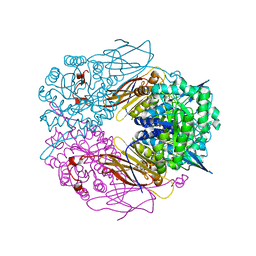 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3KT1
 
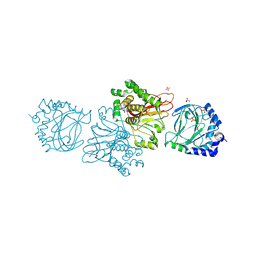 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, GLYCEROL, PKHD-type hydroxylase TPA1, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT7
 
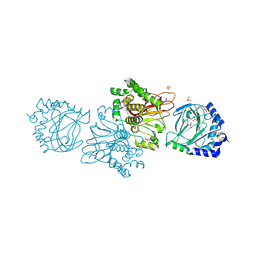 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3MYP
 
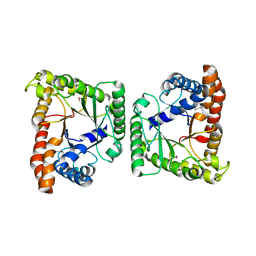 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Staphylococcus aureus | | Descriptor: | Tagatose 1,6-diphosphate aldolase | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
3NIQ
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase | | Descriptor: | 3-guanidinopropionase, GLYCEROL, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
3MYO
 
 | | Crystal structure of tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | Tagatose 1,6-diphosphate aldolase 1 | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Suh, S.W. | | Deposit date: | 2010-05-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation
Febs Lett., 585, 2011
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
