1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
3GEC
 
 | |
7POW
 
 | | Crystal structure of phosphatidyl serine synthase (PSS) in transition state. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2-amino-3-[[[(2R,3S,4R,5R)-5-(4-amino-2-oxo-pyrimidin-1-yl)-3,4-dihydroxy-tetrahydrofuran-2-yl]methoxy-hydroxy-phosphoryl]oxy-[(2R)-2,3-bis[[(Z)-octadec-9-enoyl]oxy]propoxy]-dihydroxy-lambda^5-phosphanyl]oxy-propanoic acid, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, ... | | Authors: | Yildiz, O, Centola, M. | | Deposit date: | 2021-09-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Nat Commun, 12, 2021
|
|
6F36
 
 | | Polytomella Fo model | | Descriptor: | Mitochondrial ATP synthase subunit 6, Mitochondrial ATP synthase subunit ASA6, Mitochondrial ATP synthase subunit c | | Authors: | Yildiz, O, Kuehlbrandt, W, Klusch, N, Murphy, B.J, Mills, D.J. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Elife, 6, 2017
|
|
2IWW
 
 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, beta-D-glucopyranose, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
2J9C
 
 | | Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yildiz, O, Kalthoff, C, Raunser, S, Kuehlbrandt, W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Glnk1 with Bound Effectors Indicates Regulatory Mechanism for Ammonia Uptake.
Embo J., 26, 2007
|
|
2J9D
 
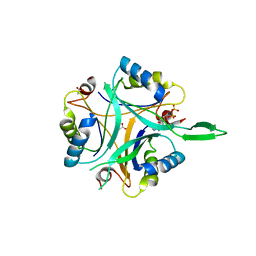 | | Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yildiz, O, Kalthoff, C, Raunser, S, Kuehlbrandt, W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Glnk1 with Bound Effectors Indicates Regulatory Mechanism for Ammonia Uptake.
Embo J., 26, 2007
|
|
2IWV
 
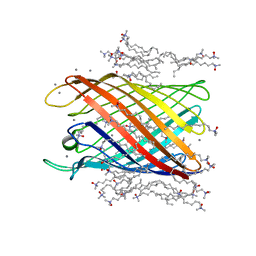 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-04 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
2J9E
 
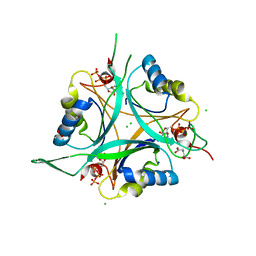 | | Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yildiz, O, Kalthoff, C, Raunser, S, Kuehlbrandt, W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Glnk1 with Bound Effectors Indicates Regulatory Mechanism for Ammonia Uptake.
Embo J., 26, 2007
|
|
7B1N
 
 | |
7B1K
 
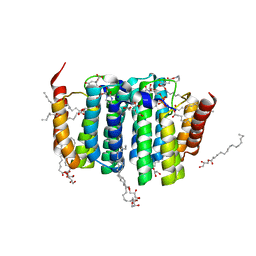 | | Crystal structure of phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CALCIUM ION, ... | | Authors: | Yildiz, O, Centola, M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Nat Commun, 12, 2021
|
|
7B1L
 
 | | Crystal structure of phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CALCIUM ION, ... | | Authors: | Yildiz, O, Centola, M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Nat Commun, 12, 2021
|
|
4V1G
 
 | |
4V1F
 
 | |
4V1H
 
 | |
2XQT
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4, DICYCLOHEXYLUREA | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
2XQS
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
2YGG
 
 | | Complex of CaMBR and CaM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Koester, S, Yildiz, O. | | Deposit date: | 2011-04-15 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structure of Human Na+/H+ Exchanger Nhe1 Regulatory Region in Complex with Cam and Ca2+
J.Biol.Chem., 286, 2011
|
|
5G3W
 
 | | Structure of HDAC like protein from Bordetella Alcaligenes in complex with the photoswitchable inhibitor CEW65 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-phenyldiazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
3DDC
 
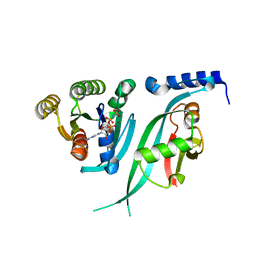 | | Crystal Structure of NORE1A in Complex with RAS | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Stieglitz, B, Bee, C, Schwarz, D, Yildiz, O, Moshnikova, A, Khokhlatchev, A, Herrmann, C. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II
Embo J., 27, 2008
|
|
5G1C
 
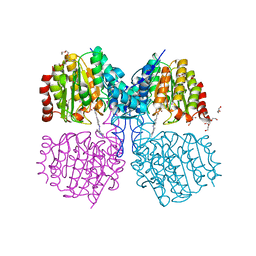 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
5G1B
 
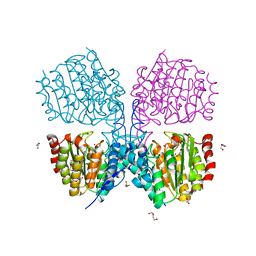 | | Bordetella Alcaligenes HDAH native | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
5G0X
 
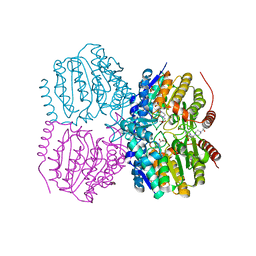 | | Pseudomonas aeruginosa HDAH bound to acetate. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, HDAH, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
5G11
 
 | | Pseudomonas aeruginosa HDAH bound to PFSAHA. | | Descriptor: | 2,2,3,3,4,4,5,5,6,6,7,7-dodecakis(fluoranyl)-~{N}-oxidanyl-~{N}'-phenyl-octanediamide, HDAH, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
5G10
 
 | | Pseudomonas aeruginosa HDAH bound to 9,9,9 trifluoro-8,8-dihydroy-N-phenylnonanamide | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HDAH, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
