4N6A
 
 | | Soybean Serine Acetyltransferase Apoenzyme | | Descriptor: | PHOSPHATE ION, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4N6B
 
 | | Soybean Serine Acetyltransferase Complexed with CoA | | Descriptor: | COENZYME A, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
3VBE
 
 | |
3VC3
 
 | | Crystal structure of beta-cyanoalanine synthase K95A mutant in soybean | | Descriptor: | N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE, beta-cyanoalnine synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Structure of Soybean beta-Cyanoalanine Synthase and the Molecular Basis for Cyanide Detoxification in Plants.
Plant Cell, 24, 2012
|
|
4N69
 
 | | Soybean Serine Acetyltransferase Complexed with Serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
3TSY
 
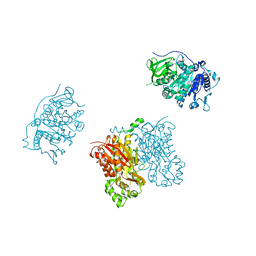 | | 4-Coumaroyl-CoA Ligase::Stilbene Synthase fusion protein | | Descriptor: | Fusion Protein 4-coumarate--CoA ligase 1, Resveratrol synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Kinetic Analysis of the Unnatural Fusion Protein 4-Coumaroyl-CoA Ligase::Stilbene Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
5XJU
 
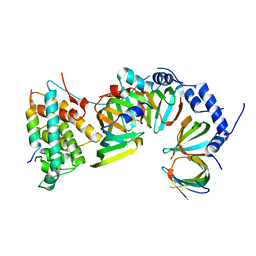 | |
5XJR
 
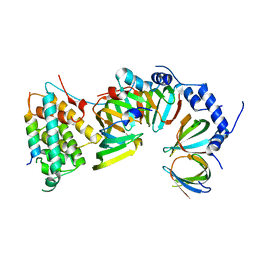 | |
5XJQ
 
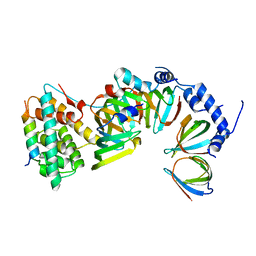 | |
5XJS
 
 | |
5XJT
 
 | |
2JRV
 
 | | The third dimensional structure of mab198-bound pep.1 for autoimmune myasthenia gravis | | Descriptor: | PEPTIDE PEP.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2JRW
 
 | | Solution structure of Cyclic extended Pep1(Cyc.ext.Pep.1) for autoimmune myasthenia gravis | | Descriptor: | Cyclic extended Pep.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2K4U
 
 | | Solution structure of the SCORPION TOXIN ADWX-1 | | Descriptor: | Potassium channel toxin alpha-KTx 3.6 | | Authors: | Yin, S.J, Jiang, L, Yi, H, Han, S, Yang, D.W, Liu, M.L, Liu, H, Cao, Z.J, Wu, Y.L, Li, W.X. | | Deposit date: | 2008-06-18 | | Release date: | 2008-12-09 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Different Residues in Channel Turret Determining the Selectivity of ADWX-1 Inhibitor Peptide between Kv1.1 and Kv1.3 Channels
J.Proteome Res., 7, 2008
|
|
5GLD
 
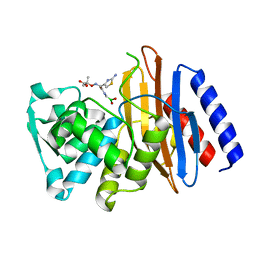 | | Crystal structure of the class A beta-lactamase PenL-tTR11 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLA
 
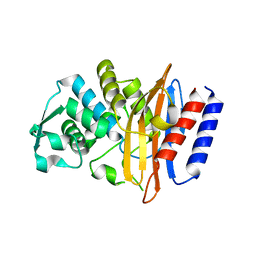 | | Crystal structure of the class A beta-lactamase PenL-tTR10 containing 10 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GL9
 
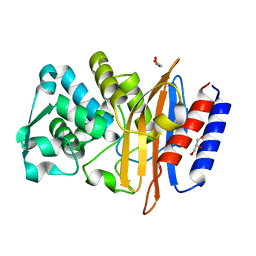 | | Crystal structure of the class A beta-lactamase PenL | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLB
 
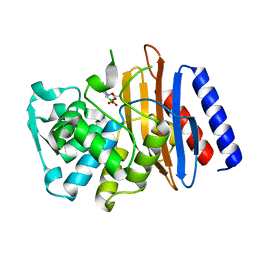 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLC
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 containing 20 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5YMY
 
 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
