4MBQ
 
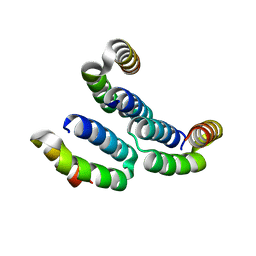 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
3JZZ
 
 | | Crystal structure of Pseudomonas aeruginosa (strain: Pa110594) typeIV pilin in space group P212121 | | Descriptor: | Type IV pilin structural subunit | | Authors: | Nguyen, Y, Jackson, S.G, Aidoo, F, Junop, M.S, Burrows, L.L. | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural characterization of Novel Pseudomonas aeruginosa type IV pilins.
J.Mol.Biol., 395, 2010
|
|
4IPV
 
 | |
4IPU
 
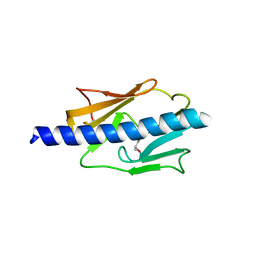 | |
1T4W
 
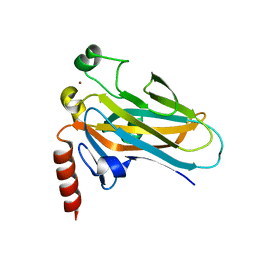 | | Structural Differences in the DNA Binding Domains of Human p53 and its C. elegans Ortholog Cep-1: Structure of C. elegans Cep-1 | | Descriptor: | C.Elegans p53 tumor suppressor-like transcription factor, ZINC ION | | Authors: | Huyen, Y, Jeffrey, P.D, Derry, W.B, Rothman, J.H, Pavletich, N.P, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Differences in the DNA Binding Domains of Human p53 and Its C. elegans Ortholog Cep-1.
Structure, 12, 2004
|
|
1XNI
 
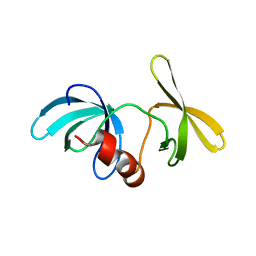 | | Tandem Tudor Domain of 53BP1 | | Descriptor: | Tumor suppressor p53-binding protein 1 | | Authors: | Huyen, Y, Zgheib, O, DiTullio Jr, R.A, Gorgoulis, V.G, Zacharatos, P, Petty, T.J, Sheston, E.A, Mellert, H.S, Stavridi, E.S, Halazonetis, T.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks
Nature, 432, 2004
|
|
3JYZ
 
 | | Crystal structure of Pseudomonas aeruginosa (strain: Pa110594) typeIV pilin in space group P41212 | | Descriptor: | SULFATE ION, Type IV pilin structural subunit | | Authors: | Nguyen, Y, Jackson, S.G, Aidoo, F, Junop, M.S, Burrows, L.L. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of Novel Pseudomonas aeruginosa type IV pilins.
J.Mol.Biol., 395, 2010
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4NOA
 
 | |
3HF1
 
 | | Crystal structure of human p53R2 | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase subunit M2 B, SULFATE ION | | Authors: | Smith, P, Zhou, B, Yuan, Y.-C, Su, L, Tsai, S.-C, Yen, Y. | | Deposit date: | 2009-05-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A X-ray crystal structure of human p53R2, a p53-inducible ribonucleotide reductase .
Biochemistry, 48, 2009
|
|
4Y15
 
 | | SdiA in complex with 3-oxo-C6-homoserine lactone | | Descriptor: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, SULFATE ION, Transcriptional regulator of ftsQAZ gene cluster | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
4Y13
 
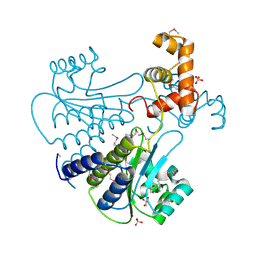 | | SdiA in complex with octanoyl-rac-glycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, GLYCEROL, SULFATE ION, ... | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
6QG3
 
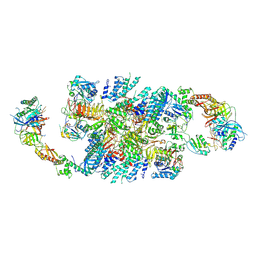 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model B) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG5
 
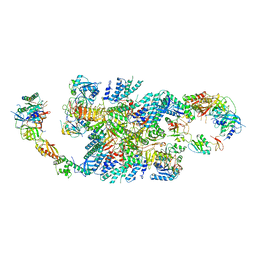 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
5MDW
 
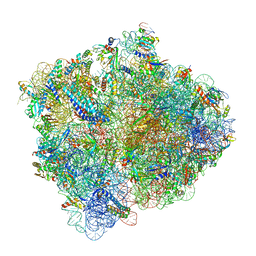 | | Structure of ArfA(A18T) and RF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
4Y17
 
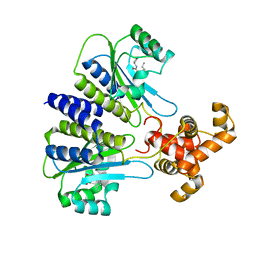 | | SdiA in complex with 3-oxo-C8-homoserine lactone | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, Transcriptional regulator of ftsQAZ gene cluster | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
5MDZ
 
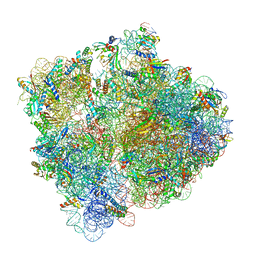 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDY
 
 | | Structure of ArfA and TtRF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDV
 
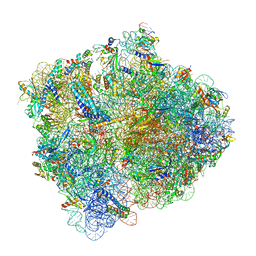 | | Structure of ArfA and RF2 bound to the 70S ribosome (accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
6BBK
 
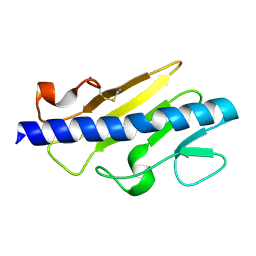 | |
4BKX
 
 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
4CYI
 
 | | Chaetomium thermophilum Pan3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
3D36
 
 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
4CYK
 
 | | Structural basis for binding of Pan3 to Pan2 and its function in mRNA recruitment and deadenylation | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3, ZINC ION | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation.
Embo J., 33, 2014
|
|
