6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UCX
 
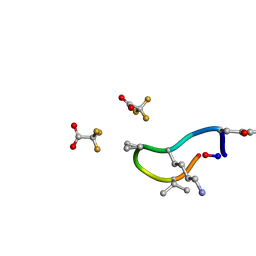 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
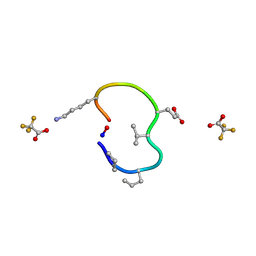 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UD9
 
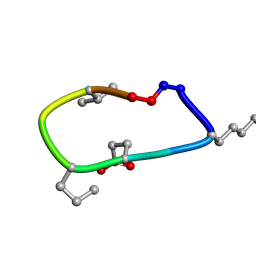 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
8G3K
 
 | |
8G4H
 
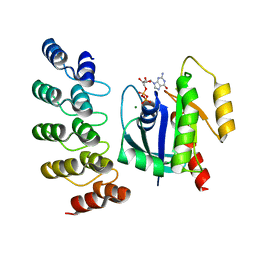 | | KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G47
 
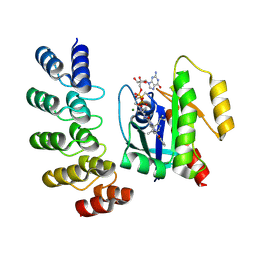 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4E
 
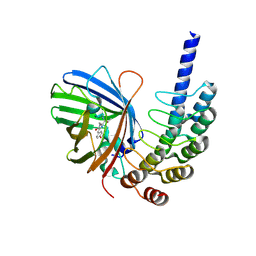 | |
8G42
 
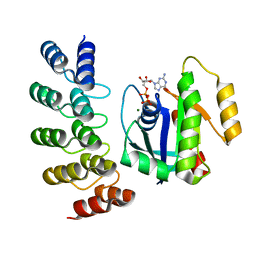 | | KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4F
 
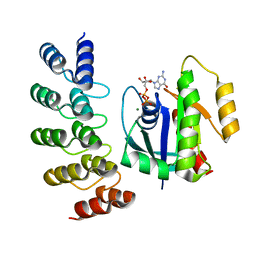 | | KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6NHT
 
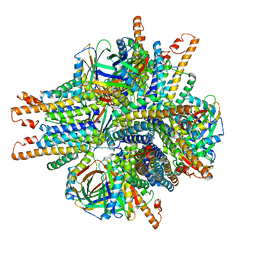 | |
6NHV
 
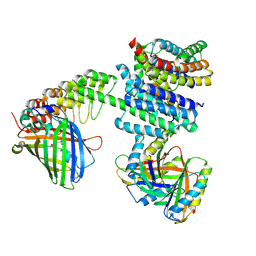 | |
8UJA
 
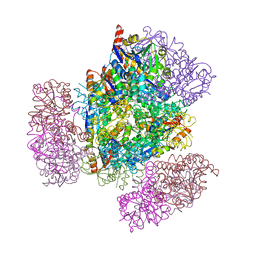 | |
1KIB
 
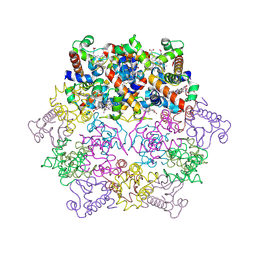 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4EGG
 
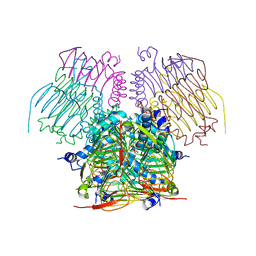 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | Descriptor: | GLYCEROL, Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
2QW7
 
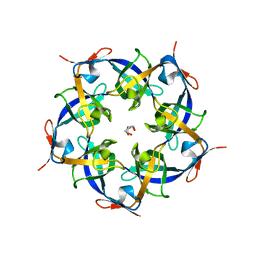 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
4FDZ
 
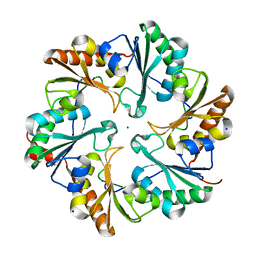 | | EutL from Clostridium perfringens, Crystallized Under Reducing Conditions | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
2RCF
 
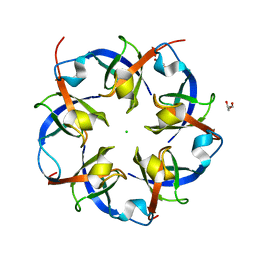 | | Carboxysome Shell protein, OrfA from H. Neapolitanus | | Descriptor: | CHLORIDE ION, GLYCEROL, Unidentified carboxysome polypeptide | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2RIF
 
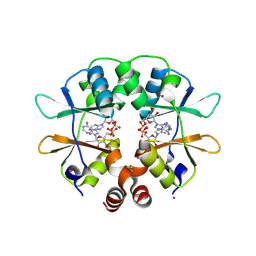 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
2RIH
 
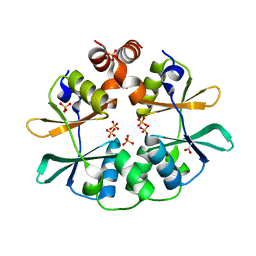 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, Conserved protein with 2 CBS domains, SULFATE ION | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon
J.Mol.Biol., 380, 2008
|
|
4EDI
 
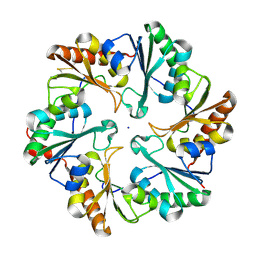 | | Disulfide bonded EutL from Clostridium perfringens | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
8UMP
 
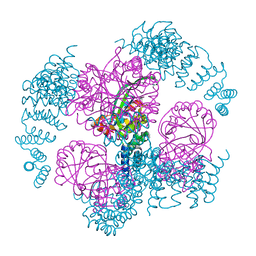 | | T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml35-redesigned-CutA-fold, T33-ml35-redesigned-TPR-domain-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UF0
 
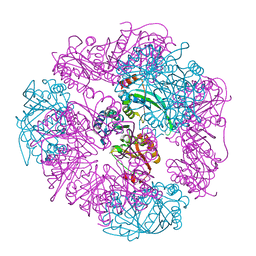 | | T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml23-redesigned-CutA-fold, T33-ml23-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-03 | | Release date: | 2023-11-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UI2
 
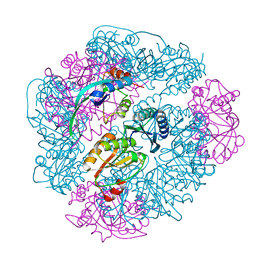 | | T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml28-redesigned-CutA-fold, T33-ml28-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UKM
 
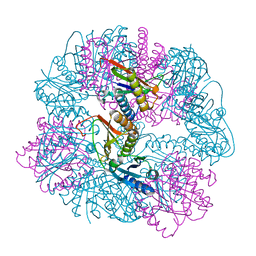 | | T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml30-redesigned-4-OT-fold, T33-ml30-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
