6XL5
 
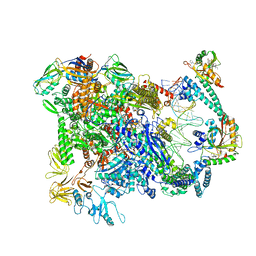 | | Cryo-EM structure of EcmrR-RNAP-promoter open complex (EcmrR-RPo) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLN
 
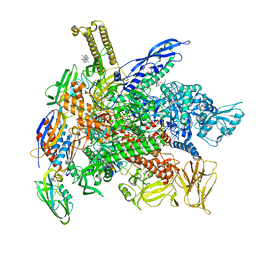 | | Cryo-EM structure of E. coli RNAP-DNA elongation complex 2 (RDe2) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLM
 
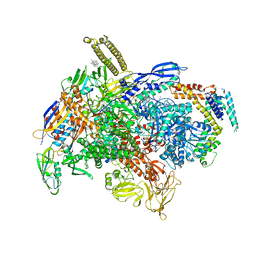 | | Cryo-EM structure of E.coli RNAP-DNA elongation complex 1 (RDe1) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLL
 
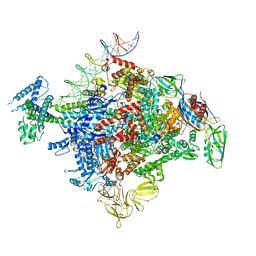 | | Cryo-EM structure of E. coli RNAP-promoter initial transcribing complex with 5-nt RNA transcript (RPitc-5nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
1MYO
 
 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, 44 STRUCTURES | | Descriptor: | MYOTROPHIN | | Authors: | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
1L1C
 
 | | Structure of the LicT Bacterial Antiterminator Protein in Complex with its RNA Target | | Descriptor: | Transcription antiterminator licT, licT mRNA antiterminator hairpin | | Authors: | Yang, Y, Declerck, N, Manival, X, Aymerich, S, Kochoyan, M. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT.
EMBO J., 21, 2002
|
|
2EC6
 
 | | Placopecten Striated Muscle Myosin II | | Descriptor: | CALCIUM ION, Myosin essential light chain, Myosin heavy chain, ... | | Authors: | Yang, Y, Brown, J, Samudrala, G, Reutzel, R, Szent-Gyorgyi, A. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
6WVD
 
 | | Human JAGN1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Green fluorescent protein, Protein jagunal homolog 1 chimera | | Authors: | Yang, Y, Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
6OC4
 
 | | CSP1-cyc(Dab6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OBW
 
 | | CSP1-cyc(K6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OC2
 
 | | CSP1-cyc(Orn6D10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V1N
 
 | | CSP1-E1A-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1FJN
 
 | |
5KJ5
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLL
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived tautomeric intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | (3~{E},5~{E})-6-oxidanyl-2-oxidanylidene-hexa-3,5-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLO
 
 | | Crystal structure of thioacyl intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169A | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLN
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169A in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLK
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ and 2-hydroxymuconate-6-semialdehyde | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
6MGS
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase with Space Group of C2221 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Davis, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.131 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6U6F
 
 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
3I5F
 
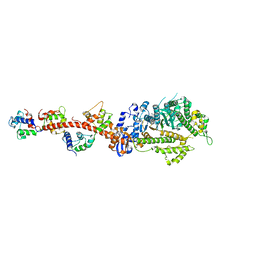 | | Crystal structure of squid MG.ADP myosin S1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
1N5H
 
 | | Solution structure of the cathelin-like domain of protegrins (the R87-P88 and D118-P119 amide bonds are in the cis conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
1N5P
 
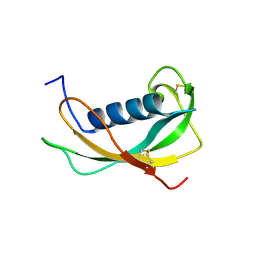 | | Solution structure of the cathelin-like domain of protegrins (all amide bonds involving proline residues are in trans conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
3I5I
 
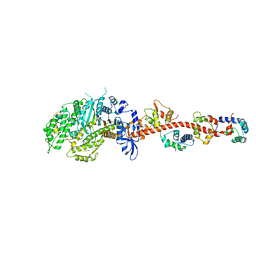 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
