4Y00
 
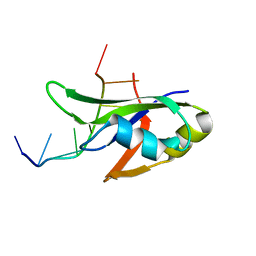 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
1K6Y
 
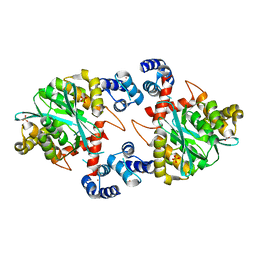 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
1K99
 
 | |
8EBT
 
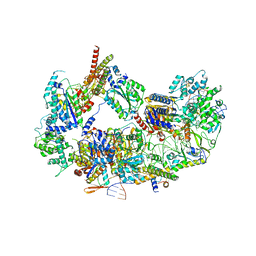 | |
8EBS
 
 | |
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBX
 
 | |
8EBW
 
 | |
8EBU
 
 | | XPC release from Core7-XPA-DNA (Cy5) | | Descriptor: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBV
 
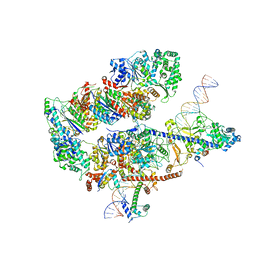 | |
1JXL
 
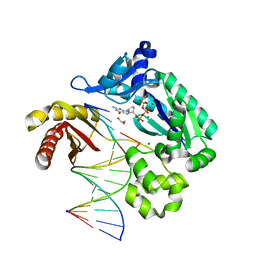 | | Crystal Structure of a Y-Family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 1,2-ETHANEDIOL, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*T)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-07 | | Release date: | 2001-10-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1JX4
 
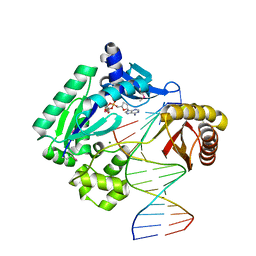 | | Crystal Structure of a Y-family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*TP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-05 | | Release date: | 2001-10-05 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1LWW
 
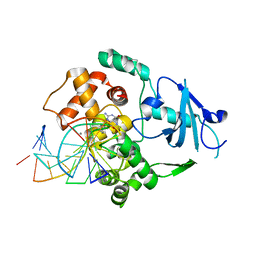 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-bromoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-BROMOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1LWV
 
 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-aminoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-AMINOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1LWY
 
 | | hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE DNA GLYCOSYLASE | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1NHJ
 
 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | Descriptor: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1NHH
 
 | | Crystal structure of N-terminal 40KD MutL protein (LN40) complex with ADPnP and one Rubidium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1ETX
 
 | |
1ETY
 
 | |
1EWR
 
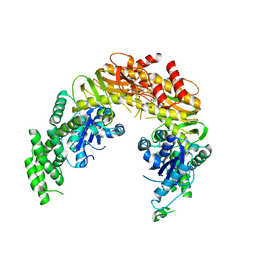 | | CRYSTAL STRUCTURE OF TAQ MUTS | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
3BSU
 
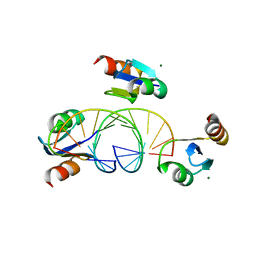 | | Hybrid-binding domain of human RNase H1 in complex with 12-mer RNA/DNA | | Descriptor: | DNA (5'-D(*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*(5IU)P*DGP*DTP*DC)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3'), ... | | Authors: | Nowotny, M, Cerritelli, S.M, Ghirlando, R, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2007-12-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD.
Embo J., 27, 2008
|
|
1ETO
 
 | |
1EA6
 
 | |
1ETQ
 
 | |
1EWQ
 
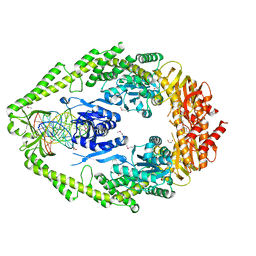 | | CRYSTAL STRUCTURE TAQ MUTS COMPLEXED WITH A HETERODUPLEX DNA AT 2.2 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3'), ... | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
