5ZYT
 
 | |
5ZYU
 
 | | The crystal structure of humanMGME1 with single strand DNA2 | | Descriptor: | DNA (5'-D(P*CP*AP*AP*CP*AP*AP*CP*A)-3'), GLYCEROL, Mitochondrial genome maintenance exonuclease 1 | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural insights into DNA degradation by human mitochondrial nuclease MGME1
Nucleic Acids Res., 46, 2018
|
|
7MEI
 
 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MKA
 
 | | Structure of EC+EC (leading EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
 | | Complex structure of trailing EC of EC+EC (trailing EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML0
 
 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML3
 
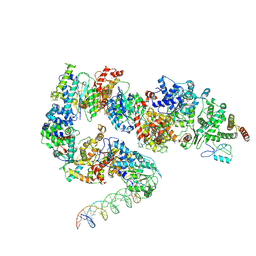 | | General transcription factor TFIIH (weak binding) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, General transcription and DNA repair factor IIH, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
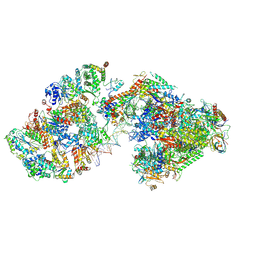 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7CKK
 
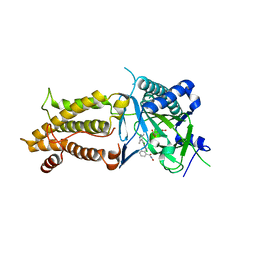 | | Structural complex of FTO bound with Dac51 | | Descriptor: | 2-{[2,6-dichloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino}-N-hydroxybenzamide, Alpha-ketoglutarate-dependent dioxygenase FTO, N-OXALYLGLYCINE | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance.
Cell Metab., 33, 2021
|
|
3J17
 
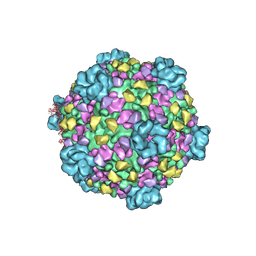 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7BPB
 
 | | Human AAA+ ATPase VCP mutant - T76E, AMP-PNP bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-22 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP9
 
 | | Human AAA+ ATPase VCP mutant - T76E, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BPA
 
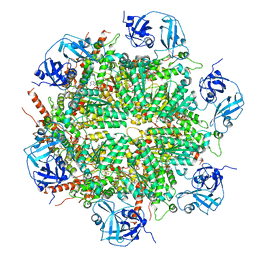 | | Human AAA+ ATPase VCP mutant - T76A, AMP-PNP-bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP8
 
 | | Human AAA+ ATPase VCP mutant - T76A, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
3CE7
 
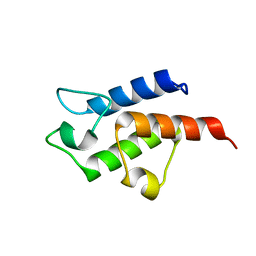 | | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510 | | Descriptor: | Specific mitochondrial acyl carrier protein | | Authors: | Wernimont, A.K, Dong, A, Yang, C, Khuu, C, Lam, A, Brand, V, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510.
To be Published
|
|
3KCW
 
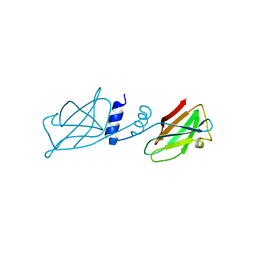 | | Crystal structure of Ganoderma fungal immunomodulatory protein, GMI | | Descriptor: | immunomodulatory protein | | Authors: | Hsu, M.F, Wang, A.H.J, Yang, C.S, Huang, C.T, Hseu, R.S, Lin, C.W, Wu, M.Y, Huang, C.S, Fu, H.Y. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single cysteine replacement at Leu6 increase the potent and thermostability in Ganoderma fungal immunomodulatory proteins
To be Published
|
|
4WAV
 
 | | Crystal Structure of Haloquadratum walsbyi bacteriorhodopsin mutant D93N | | Descriptor: | Bacteriorhodopsin-I, RETINAL, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Wang, A.H.J, Hsu, M.F, Yang, C.S, Fu, H.Y. | | Deposit date: | 2014-09-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an acid-tolerant light-driven proton pump at 1.85 Angstroms resolution
To be published
|
|
6QVJ
 
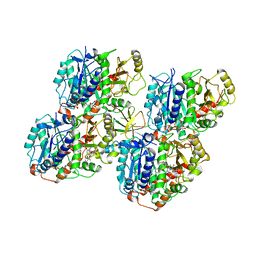 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QVE
 
 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
6QUS
 
 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
1BQJ
 
 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
9M0Q
 
 | |
