6DOT
 
 | |
6DOJ
 
 | |
6DMV
 
 | |
6DOI
 
 | |
1NHI
 
 | | Crystal structure of N-terminal 40KD MutL (LN40) complex with ADPnP and one potassium | | 分子名称: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | 著者 | Hu, X, Machius, M, Yang, W. | | 登録日 | 2002-12-19 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
5L9X
 
 | |
5C8Z
 
 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | 分子名称: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | 著者 | Hu, X.-J, Qi, Q, Yang, W.-J. | | 登録日 | 2015-06-26 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4BT0
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-12 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DXY
 
 | | Crystal structures of CYP101D2 Y96A mutant | | 分子名称: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhou, W, Bell, S.G, Yang, W, Dale, A, Wong, L.-L. | | 登録日 | 2012-02-28 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Improving the affinity and activity of CYP101D2 for hydrophobic substrates
Appl.Microbiol.Biotechnol., 2012
|
|
4BT1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-12 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6DMN
 
 | |
4DNJ
 
 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | 分子名称: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | 登録日 | 2012-02-08 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4BS1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-06 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | 分子名称: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | 登録日 | 2010-08-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3OFT
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | 分子名称: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | 登録日 | 2010-08-16 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
4EGP
 
 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | 分子名称: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | 著者 | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | 登録日 | 2012-03-31 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
1Q2O
 
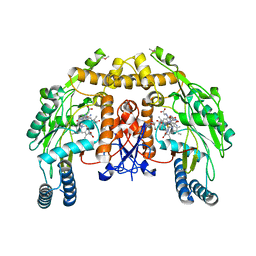 | | Bovine endothelial nitric oxide synthase N368D mutant heme domain dimer with L-N(omega)-nitroarginine-2,4-L-diaminobutyramide bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | 著者 | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | 登録日 | 2003-07-25 | | 公開日 | 2004-01-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6OEO
 
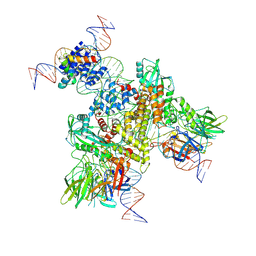 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEQ
 
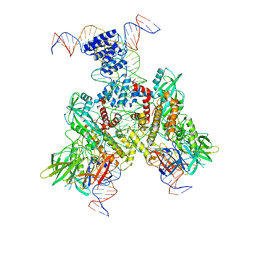 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OER
 
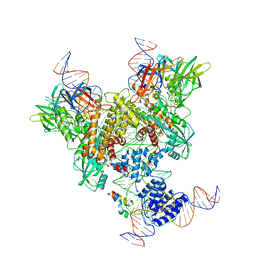 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OES
 
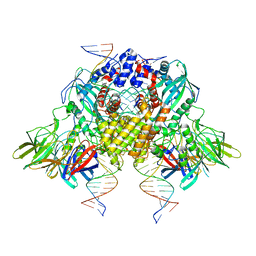 | | Cryo-EM structure of mouse RAG1/2 STC complex (without NBD domain) | | 分子名称: | CALCIUM ION, DNA (34-MER), DNA (35-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEM
 
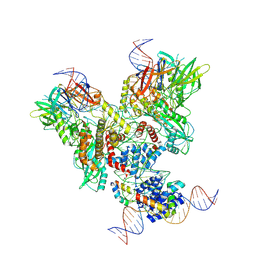 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | 分子名称: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
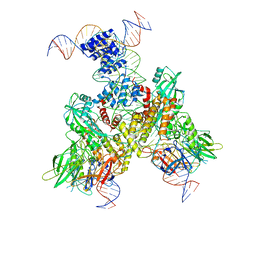 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | 分子名称: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
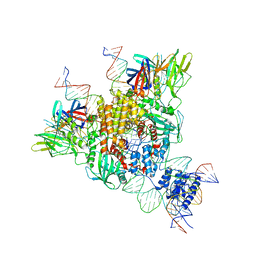 | | Cryo-EM structure of mouse RAG1/2 STC complex | | 分子名称: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | 著者 | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | 登録日 | 2019-03-27 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
