5XBZ
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
6LIG
 
 | |
5Z9S
 
 | |
2M02
 
 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
2MPX
 
 | |
3E8N
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) complexed with a potent inhibitor RDEA119 and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Yan, S, Wang, Z.M. | | Deposit date: | 2008-08-20 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RDEA119/BAY 869766: a potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer.
Cancer Res., 69, 2009
|
|
2IJN
 
 | | Isothiazoles as active-site inhibitors of HCV NS5B polymerase | | Descriptor: | (2R,3R)-3-{[3,5-BIS(TRIFLUOROMETHYL)PHENYL]AMINO}-2-CYANO-3-THIOXOPROPANAMIDE, RNA polymerase NS5B | | Authors: | Yan, S, Yao, N. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazoles as active-site inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5J76
 
 | | Structure of Lectin from Colocasia esculenta(L.) Schott | | Descriptor: | 12kD storage protein, GLYCEROL | | Authors: | Vajravijayan, S, Pletnev, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-04-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of beta-prism lectin from Colocasia esculenta (L.) S chott.
Int.J.Biol.Macromol., 91, 2016
|
|
1GQ4
 
 | |
1GQ5
 
 | |
5WQU
 
 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | Authors: | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | Deposit date: | 2017-10-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
5WQS
 
 | | Crystal structure of Apo Beta-Amylase from Sweet potato | | Descriptor: | Beta-amylase, ISOPROPYL ALCOHOL | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
6LIK
 
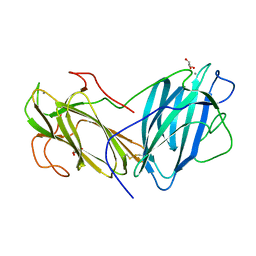 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with Galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Iqbal, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6VNW
 
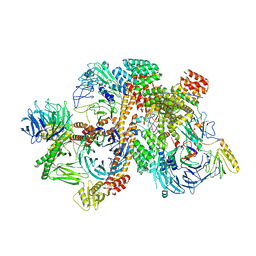 | | Cryo-EM structure of apo-BBSome | | Descriptor: | BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, Bardet-Biedl syndrome 2 protein homolog, ... | | Authors: | Yang, S, Walz, T, Nachury, M, Chou, H. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
6VOA
 
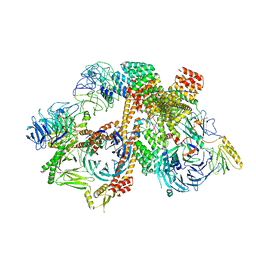 | | Cryo-EM structure of the BBSome-ARL6 complex | | Descriptor: | ADP-ribosylation factor-like protein 6, BBS1 domain-containing protein, Bardet-Biedl syndrome 18 protein, ... | | Authors: | Yang, S, Walz, T, Nachury, M.V. | | Deposit date: | 2020-01-30 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes.
Elife, 9, 2020
|
|
6LI7
 
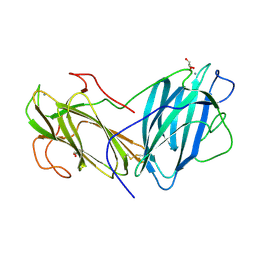 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Ankur, T, Gunasekaran, K, Nandhagopal, N. | | Deposit date: | 2019-12-10 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
5JJY
 
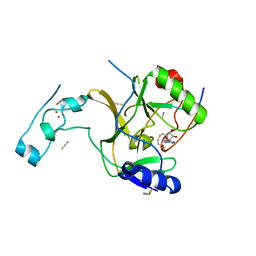 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yang, S, Zheng, X, Li, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
3L43
 
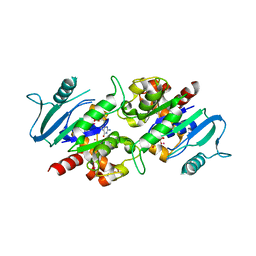 | | Crystal structure of the dynamin 3 GTPase domain bound with GDP | | Descriptor: | Dynamin-3, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Yang, S, Tempel, W, Tong, Y, Nedyalkova, L, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the dynamin 3 GTPase domain bound with GDP
to be published
|
|
8IPJ
 
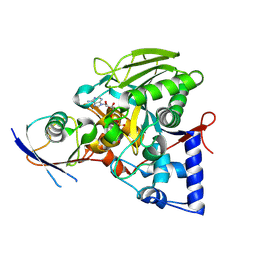 | | Crystal structure of the Legionella effector protein MavL with ADPR-Ub | | Descriptor: | MavL, Ubiquitin, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of the Legionella effector protein MavL with ADPR-Ub
To Be Published
|
|
8IPW
 
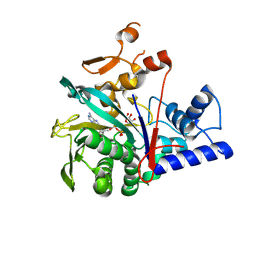 | | The sturecture of Legionella effector protein MavL with ADPR | | Descriptor: | MavL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2023-03-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The sturecture of Legionella effector protein MavL with ADPR
To Be Published
|
|
7SC5
 
 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SD3
 
 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
