6KGT
 
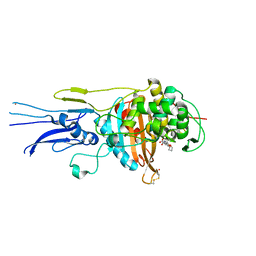 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | 分子名称: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.308 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
3O3K
 
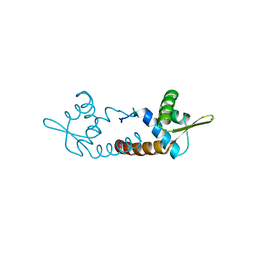 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | 分子名称: | Uncharacterized protein AF_1382 | | 著者 | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Xu, H, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2010-07-24 | | 公開日 | 2010-08-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6KGS
 
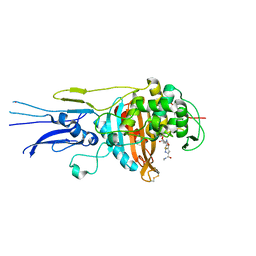 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
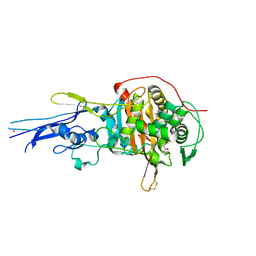 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.407 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGV
 
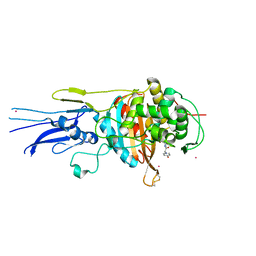 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | 著者 | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2019-07-12 | | 公開日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | 分子名称: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q62
 
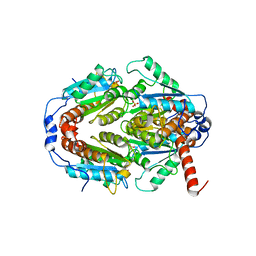 | | Crystal Structure of ArsH from Sinorhizobium meliloti | | 分子名称: | SULFATE ION, arsH | | 著者 | Ye, J, Yang, H, Bhattacharjee, H, Rosen, B.P. | | 登録日 | 2007-06-04 | | 公開日 | 2007-12-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti
FEBS Lett., 581, 2007
|
|
2Q6D
 
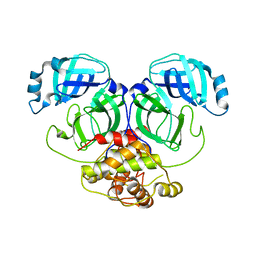 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | 分子名称: | Infectious bronchitis virus (IBV) main protease | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-04 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
8YEW
 
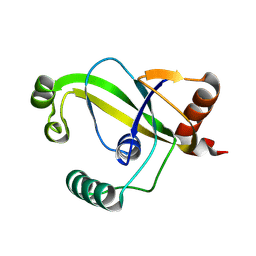 | | TRIP4 ASCH domain in unliganded form | | 分子名称: | Activating signal cointegrator 1 | | 著者 | Ding, J, Yang, H, Hu, C. | | 登録日 | 2024-02-23 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 2024
|
|
8YFI
 
 | |
8YFJ
 
 | |
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E5X
 
 | |
7WS1
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7ESX
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wPip | | 分子名称: | Bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESY
 
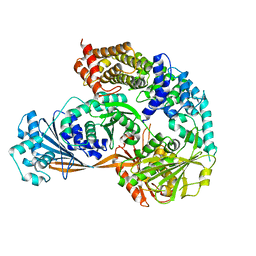 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip | | 分子名称: | Bacteria factor 1, CALCIUM ION, ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5H2C
 
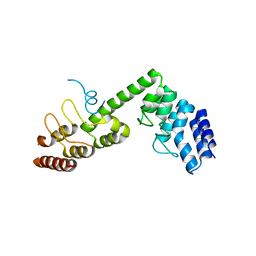 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | 分子名称: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | 著者 | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | 登録日 | 2016-10-14 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.508 Å) | | 主引用文献 | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2D
 
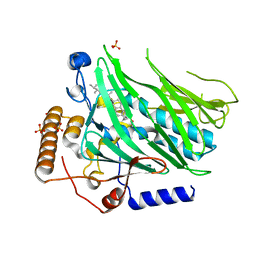 | | Crystal structure of Osh1 ORD domain in complex with ergosterol | | 分子名称: | ERGOSTEROL, KLLA0C04147p, SULFATE ION | | 著者 | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | 登録日 | 2016-10-14 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5WBU
 
 | |
7WS8
 
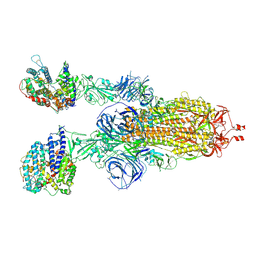 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS0
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS6
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WSA
 
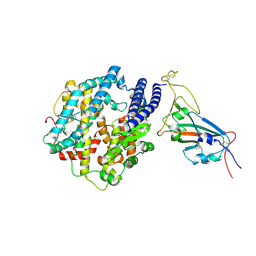 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
