5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | 分子名称: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD6
 
 | | Crystal structure of BACE complex with BMC004 | | 分子名称: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCA
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 1-{4-[(2-chloro-5-{1-[3-(4-cyclopropylpiperazin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}-N-[(4-chlorophenyl)methyl]methanamine, Cathepsin S, SULFATE ION | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QD1
 
 | | Crystal structure of BACE complex with BMC011 | | 分子名称: | (10S,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-7-oxa-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCT
 
 | | Crystal structure of BACE complex with BMC001 | | 分子名称: | (2R,4S)-N-butyl-4-[(4S,6R)-16-ethoxy-12-ethyl-6-methyl-2,13-dioxo-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-trien-4-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1, PHOSPHATE ION | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | 著者 | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | 登録日 | 2010-03-02 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
5QC7
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | 分子名称: | 2-[1-(cyclohexylmethyl)piperidin-4-yl]-1-{3-[3-{[2-(piperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-propyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, Cathepsin S, DIMETHYL SULFOXIDE, ... | | 著者 | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | 登録日 | 2017-08-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCZ
 
 | | Crystal structure of BACE complex with BMC015 | | 分子名称: | (4S)-4-{(S)-hydroxy[(3R,6R)-6-(methoxymethyl)morpholin-3-yl]methyl}-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCR
 
 | | Crystal structure of BACE complex with BMC026 | | 分子名称: | 2-(butylamino)-N-[(2S,3S,5R)-6-(butylamino)-3-hydroxy-5-methyl-6-oxo-1-phenylhexan-2-yl]-6-methoxypyridine-4-carboxamide, Beta-secretase 1, SULFATE ION | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD8
 
 | | Crystal structure of BACE complex with BMC003 | | 分子名称: | (3S,14R,16S)-16-[(1R)-2-{[(4S)-2,2-dimethyl-6-(propan-2-yl)-3,4-dihydro-2H-1-benzopyran-4-yl]amino}-1-hydroxyethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | 著者 | Ostermann, N, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | 著者 | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | 登録日 | 2015-03-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.916 Å) | | 主引用文献 | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
5QCV
 
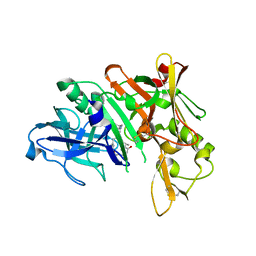 | | Crystal structure of BACE complex with BMC023 | | 分子名称: | (10S,13S)-13-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9,10-dimethyl-2-oxa-9,12-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-8,11-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDB
 
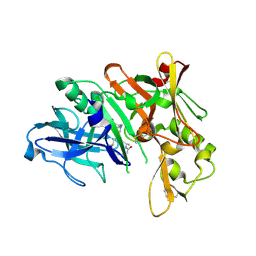 | | Crystal structure of BACE complex with BMC002 | | 分子名称: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD0
 
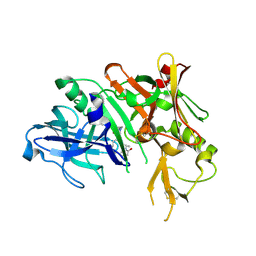 | | Crystal structure of BACE complex withBMC006 | | 分子名称: | (5S,8S,10R)-8-[(1R)-1-hydroxy-2-{[(5-propyl-1H-pyrazol-3-yl)methyl]amino}ethyl]-4,5,10-trimethyl-1-oxa-4,7-diazacyclohexadecane-3,6-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1L2Z
 
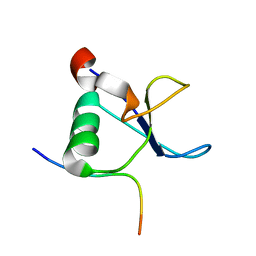 | | CD2BP2-GYF domain in complex with proline-rich CD2 tail segment peptide | | 分子名称: | CD2 ANTIGEN (CYTOPLASMIC TAIL)-BINDING PROTEIN 2, T-CELL SURFACE ANTIGEN CD2 | | 著者 | Freund, C, Kuhne, R, Yang, H, Park, S, Reinherz, E.L, Wagner, G. | | 登録日 | 2002-02-26 | | 公開日 | 2002-11-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules
Embo J., 21, 2002
|
|
4FUW
 
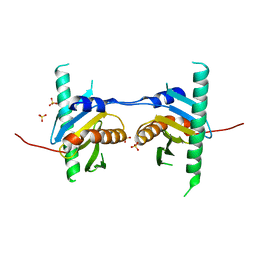 | | Crystal structure of Ego3 mutant | | 分子名称: | Protein SLM4, SULFATE ION | | 著者 | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | 登録日 | 2012-06-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
1D5R
 
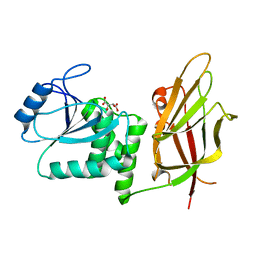 | | Crystal Structure of the PTEN Tumor Suppressor | | 分子名称: | L(+)-TARTARIC ACID, PHOSPHOINOSITIDE PHOSPHATASE PTEN | | 著者 | Lee, J.O, Yang, H, Georgescu, M.-M, Di Cristofano, A, Pavletich, N.P. | | 登録日 | 1999-10-11 | | 公開日 | 1999-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association.
Cell(Cambridge,Mass.), 99, 1999
|
|
8U78
 
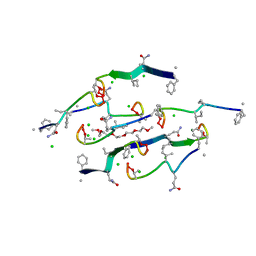 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | 著者 | Nowick, J.S, Yang, H, Kreutzer, A.G. | | 登録日 | 2023-09-14 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
3R7W
 
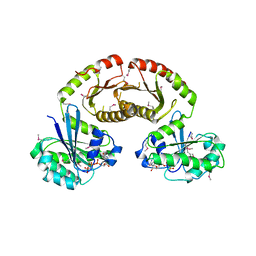 | | Crystal Structure of Gtr1p-Gtr2p complex | | 分子名称: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | 著者 | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | 登録日 | 2011-03-23 | | 公開日 | 2011-08-24 | | 最終更新日 | 2013-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.773 Å) | | 主引用文献 | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | 分子名称: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | 著者 | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | 登録日 | 2020-02-12 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
5Q0K
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0R
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,1-dibenzyl-6-[(2-fluorophenyl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 3-(2-chlorophenyl)-N-[(1R)-1-(naphthalen-2-yl)ethyl]-5-(propan-2-yl)-1,2-oxazole-4-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | 分子名称: | MP1 | | 著者 | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | 登録日 | 2003-10-28 | | 公開日 | 2005-02-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
