1YYE
 
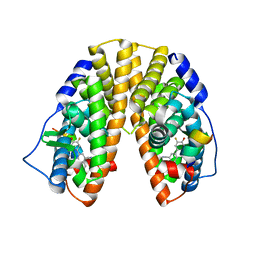 | | Crystal structure of estrogen receptor beta complexed with way-202196 | | 分子名称: | 3-(3-FLUORO-4-HYDROXYPHENYL)-7-HYDROXY-1-NAPHTHONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | 著者 | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | 登録日 | 2005-02-24 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | 分子名称: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | 著者 | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | 登録日 | 2020-03-31 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4IWG
 
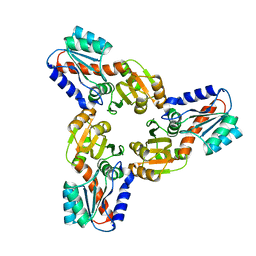 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | 分子名称: | UPF0135 protein MJ0927 | | 著者 | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | 登録日 | 2013-01-23 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.472 Å) | | 主引用文献 | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
4MG2
 
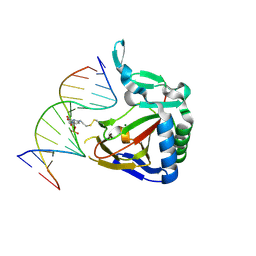 | | ALKBH2 F102A cross-linked to undamaged dsDNA | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA-1, DNA-2, ... | | 著者 | Chen, B, Gan, J, Yang, C.G. | | 登録日 | 2013-08-28 | | 公開日 | 2014-02-26 | | 最終更新日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The complex structures of ALKBH2 mutants cross-linked to
dsDNA reveal the conformational swing of β-hairpin
Sci China Chem, 57, 2014
|
|
8IMI
 
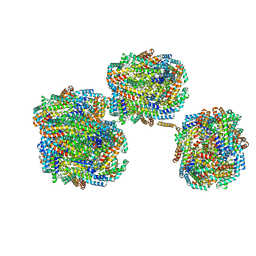 | | A1-A2, A3-A4, B'1-B'2, C'1-C'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster A) | | 分子名称: | ApcA2, ApcB2, ApcB3, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IML
 
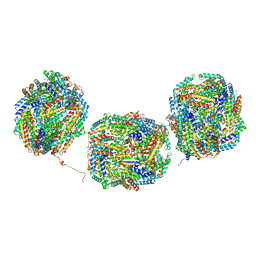 | | Rs2I-Rs2II, Rs1I-Rs1II, RbI-RbII cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster D) | | 分子名称: | CpcA, CpcB, CpcD, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMN
 
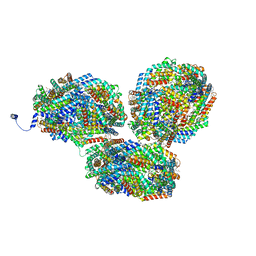 | | Rt1I-Rt1II, Rt2'I-Rt2'II, Rt3I-Rt3II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster F) | | 分子名称: | CpcA, CpcB, CpcD, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMM
 
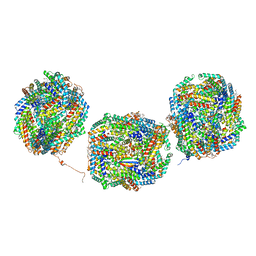 | | Rs2'I-Rs2'II, Rs1'I-Rs1'II, Rb'I-Rb'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster E) | | 分子名称: | CpcA, CpcB, CpcD, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMO
 
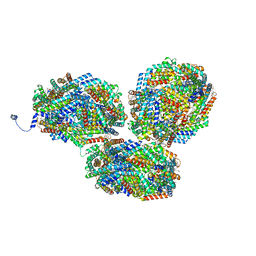 | | Rt1'I-Rt1'II, Rt2I-Rt2II, Rt3'I-Rt3'II cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster G) | | 分子名称: | CpcA, CpcB, CpcD, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
7F8T
 
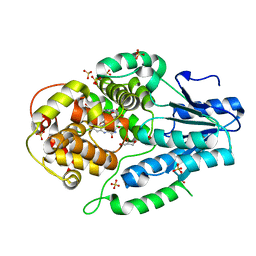 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | 分子名称: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2021-07-02 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
8IMK
 
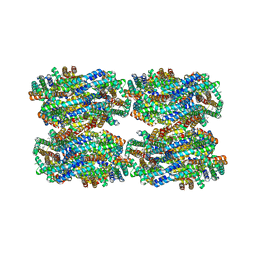 | | D3-D4, D1-D2, D'3-D'4, D'1-D'2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster C) | | 分子名称: | ApcA1, ApcA2, ApcB1, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
8IMJ
 
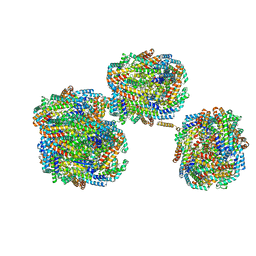 | | A'1-A'2, A'3-A'4, B1-B2, C1-C2 cylinder in cyanobacterial phycobilisome from Anthocerotibacter panamensis (Cluster B) | | 分子名称: | ApcA2, ApcB2, ApcB3, ... | | 著者 | Wang, C.H, Yang, C.H, Wu, H.Y, Jiang, H.W, Ho, M.C, Ho, M.Y. | | 登録日 | 2023-03-07 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | A structure of the relict phycobilisome from a thylakoid-free cyanobacterium.
Nat Commun, 14, 2023
|
|
2GBZ
 
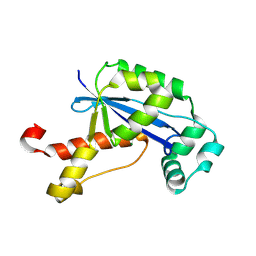 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | 分子名称: | MAGNESIUM ION, Oligoribonuclease | | 著者 | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | 登録日 | 2006-03-12 | | 公開日 | 2007-01-16 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | 分子名称: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | 著者 | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | 登録日 | 2012-03-05 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | 分子名称: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | 著者 | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | 登録日 | 2012-03-05 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | 分子名称: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | 著者 | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | 登録日 | 2012-03-05 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
8HYK
 
 | | CD-NTase EfCdnE in complex with intermediate pppU[2'-5']p | | 分子名称: | EfCdnE, MAGNESIUM ION, [[(2R,3R,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | 著者 | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2023-01-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.021 Å) | | 主引用文献 | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
8I7X
 
 | | Crystal structure of human ClpP in complex with ZG36 | | 分子名称: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | 著者 | Wang, P.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2023-02-02 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists.
Eur.J.Med.Chem., 258, 2023
|
|
8HWJ
 
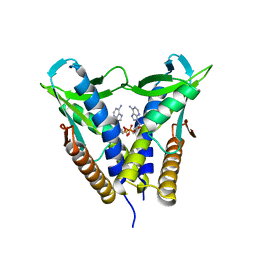 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | 著者 | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2022-12-30 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.553 Å) | | 主引用文献 | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWI
 
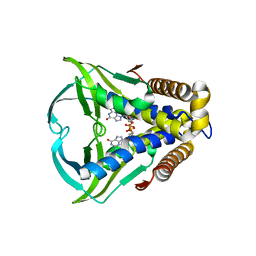 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | 著者 | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2022-12-30 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
3ODK
 
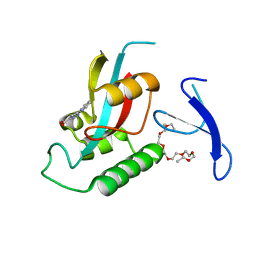 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | 登録日 | 2010-08-11 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8HY9
 
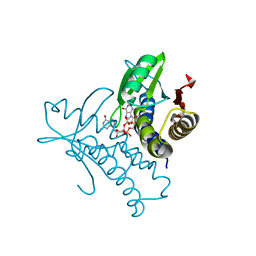 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | 著者 | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.462 Å) | | 主引用文献 | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
1NHU
 
 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | 分子名称: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | 著者 | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | 登録日 | 2002-12-19 | | 公開日 | 2003-03-18 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | 著者 | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | 登録日 | 2017-06-28 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.383 Å) | | 主引用文献 | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
4KPH
 
 | | Structure of the Fab fragment of N62, a protective monoclonal antibody to the nonreducing end of Francisella tularensis O-antigen | | 分子名称: | ACETATE ION, N62 heavy chain, N62 light chain | | 著者 | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Wang, Q, Costello, C.E, Zaia, J, Seaton, B.A, Sharon, J. | | 登録日 | 2013-05-13 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | The binding sites of monoclonal antibodies to the non-reducing end of Francisella tularensis O-antigen accommodate mainly the terminal saccharide.
Immunology, 140, 2013
|
|
