6ZAR
 
 | | As-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.1 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAW
 
 | | Damage-free NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAX
 
 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at low dose (0.5 MGy) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAU
 
 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAS
 
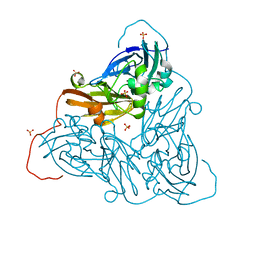 | | Damage-free as-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZAT
 
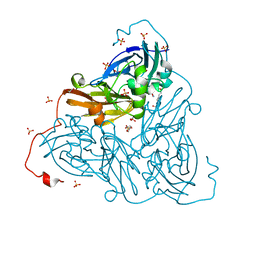 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.0 A resolution (unrestrained full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
8ILK
 
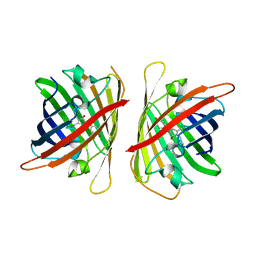 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
5ONX
 
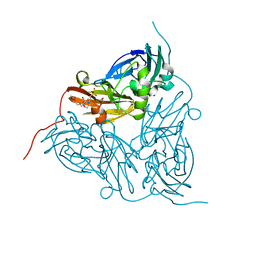 | | Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5ONY
 
 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
6LLQ
 
 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8J81
 
 | | MDM2 bound with a peptoid | | Descriptor: | (2S)-2-[[(2S)-2-[(6-chloranyl-1H-indol-3-yl)methyl-[(2S)-2-[[(2S)-2-[ethanoyl-(phenylmethyl)amino]propanoyl]-methyl-amino]propanoyl]amino]propanoyl]-methyl-amino]-N-(3,3-dimethylbutyl)-N-[(2S)-1-oxidanylidene-1-piperazin-1-yl-propan-2-yl]propanamide, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yokomine, M, Fukuda, Y, Ago, H, Matsuura, H, Ueno, G, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Jumpei, M, Sando, S. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structural and physicochemical study of how a peptoid binds to a protein
To Be Published
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6GSQ
 
 | | Oxidised copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GT2
 
 | | Reduced copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GT0
 
 | | Nitrite-bound copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6M5E
 
 | | Human serum albumin with cyclic peptide dalbavancin | | Descriptor: | (2S,5S)-5-azanyl-3,4,6-tris(oxidanyl)oxane-2-carboxylic acid, 10-METHYLUNDECANOIC ACID, 2-amino-2-deoxy-beta-D-altropyranuronic acid, ... | | Authors: | Ito, S, Senoo, A, Nagatoishi, S, Ohue, M, Yamamoto, M, Tsumoto, K, Wakui, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Mechanism of Human Serum Albumin Complexed with Cyclic Peptide Dalbavancin.
J.Med.Chem., 63, 2020
|
|
1X1S
 
 | | Crystal structure of M-Ras in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
1X1R
 
 | | Crystal structure of M-Ras in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein M-Ras | | Authors: | Ye, M, Shima, F, Muraoka, S, Liao, J, Okamoto, H, Yamamoto, M, Tamura, A, Yagi, N, Ueki, T, Kataoka, T. | | Deposit date: | 2005-04-12 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of M-Ras Reveals a GTP-bound "Off" State Conformation of Ras Family Small GTPases
J.Biol.Chem., 280, 2005
|
|
1X2R
 
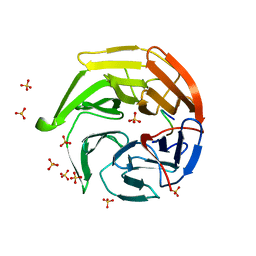 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nuclear factor erythroid 2 related factor 2, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
1WN6
 
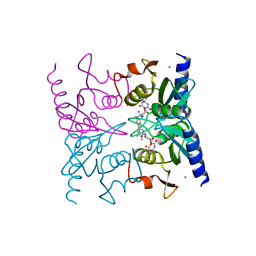 | | Crystal Structure of Blasticidin S Deaminase (BSD) Complexed with Tetrahedral Intermediate of Blasticidin S | | Descriptor: | 6-(4-AMINO-4-HYDROXY-2-OXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-3-[3-AMINO-5-(N-METHYL-GUANIDINO)-PENT ANOYLAMINO]-3,6-DIHYDRO-2H-PYRAN-2-CARBOXYLIC ACID, ARSENIC, Blasticidin-S deaminase, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
4EFM
 
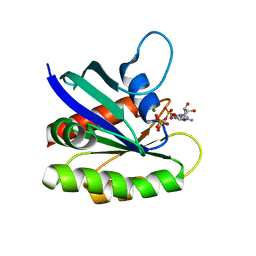 | | Crystal structure of H-Ras G12V in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
