7XI9
 
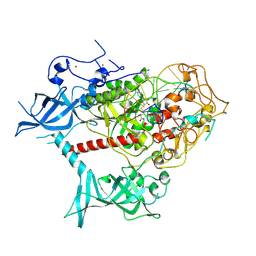 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
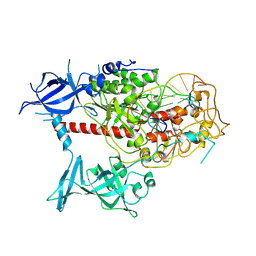 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
3W0W
 
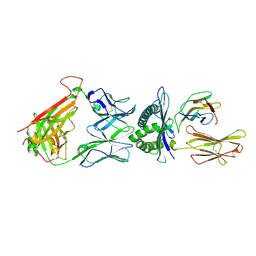 | | The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide in space group P212121 | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXS
 
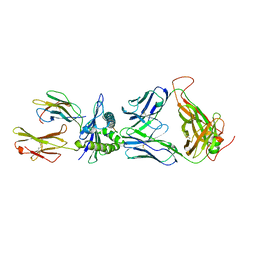 | | The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(6L) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, H27-14 TCR alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXR
 
 | | The complex between H27-14 TCR and HLA-A24 bound to HIV-1 Nef134-10(wt) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, H27-14 TCR alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXP
 
 | | HLA-A24 in complex with HIV-1 Nef134-10(6L) | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXN
 
 | | HLA-A24 in complex with HIV-1 Nef134-10(wt) | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXT
 
 | | T36-5 TCR specific for HLA-A24-Nef134-10 | | Descriptor: | T36-5 TCR alpha chain, T36-5 TCR beta chain | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXO
 
 | | HLA-A24 in complex with HIV-1 Nef134-10(2F) | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXU
 
 | | The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXM
 
 | | The complex between C1-28 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, C1-28 TCR alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
3VXQ
 
 | | H27-14 TCR specific for HLA-A24-Nef134-10 | | Descriptor: | H27-14 TCR alpha chain, H27-14 TCR beta chain | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
6IXF
 
 | |
6IXE
 
 | | Crystal structure of SeMet apo SH3BP5 (I41) | | Descriptor: | SH3 domain-binding protein 5, SUCCINIC ACID | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2018-12-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6IXV
 
 | | Crystal structure of SH3BP5-Rab11a | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-11A, SH3 domain-binding protein 5 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6IXG
 
 | |
3A58
 
 | | Crystal structure of Sec3p - Rho1p complex from Saccharomyces cerevisiae | | Descriptor: | Exocyst complex component SEC3, GTP-binding protein RHO1, MAGNESIUM ION, ... | | Authors: | Yamashita, M, Sato, Y, Yamagata, A, Mimura, H, Yoshikawa, A, Fukai, S. | | Deposit date: | 2009-08-03 | | Release date: | 2010-01-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3
Nat.Struct.Mol.Biol., 17, 2010
|
|
5GVC
 
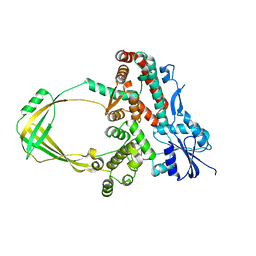 | | Human Topoisomerase IIIb topo domain | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVD
 
 | | Human TDRD3 DUF1767-OB domains | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVE
 
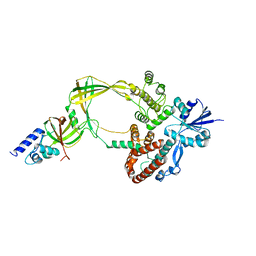 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5H11
 
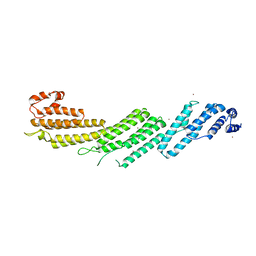 | |
3A9J
 
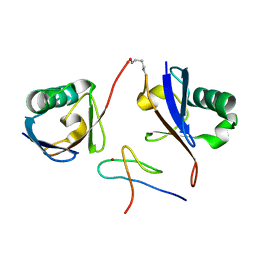 | | Crystal structure of the mouse TAB2-NZF in complex with Lys63-linked di-ubiquitin | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2, Ubiquitin, ZINC ION | | Authors: | Sato, Y, Yoshikawa, A, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3
Embo J., 28, 2009
|
|
3A1Q
 
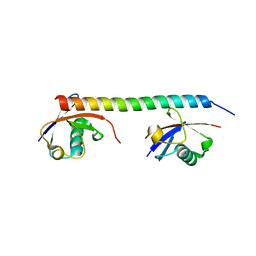 | | Crystal structure of the mouse RAP80 UIMs in complex with Lys63-linked di-ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin interaction motif-containing protein 1 | | Authors: | Sato, Y, Yoshikawa, A, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80
Embo J., 28, 2009
|
|
3B08
 
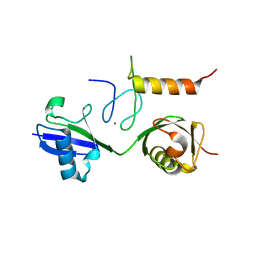 | | Crystal structure of the mouse HOIL1-L-NZF in complex with linear di-ubiquitin | | Descriptor: | Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION, ... | | Authors: | Sato, Y, Fujita, H, Yoshikawa, A, Yamashita, M, Yamagata, A, Kaiser, S.E, Iwai, K, Fukai, S. | | Deposit date: | 2011-06-07 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ZNV
 
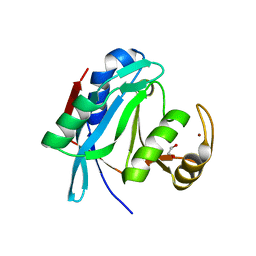 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
