3K7R
 
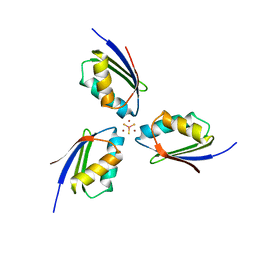 | | Crystal structure of [TM][CuAtx1]3 | | Descriptor: | COPPER (II) ION, D-MALATE, Metal homeostasis factor ATX1, ... | | Authors: | Xue, Y, Alvarez, H.M, Robinson, C.D, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation.
Science, 327, 2010
|
|
4ACH
 
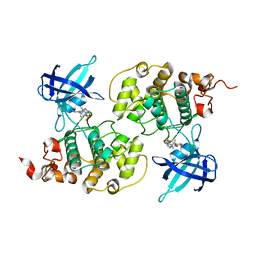 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-N-(3-METHOXYPROPYL)-6-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}PYRAZINE-2-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
4ACD
 
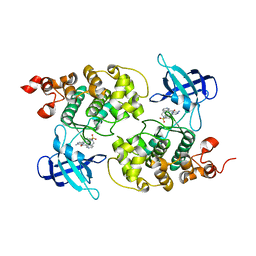 | | GSK3b in complex with inhibitor | | Descriptor: | 3-AMINO-6-{4-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-N-PYRIDIN-3-YLPYRAZINE-2-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Xue, Y, Ormo, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel potent and highly selective glycogen synthase kinase-3 beta (GSK3 beta ) inhibitors for Alzheimer's disease: design, synthesis, and characterization of pyrazines.
J. Med. Chem., 55, 2012
|
|
2AFV
 
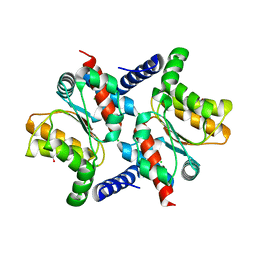 | | The Crystal Structure of Putative Precorrin Isomerase CbiC in Cobalamin Biosynthesis | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Xue, Y, Wei, Z. | | Deposit date: | 2005-07-26 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of putative precorrin isomerase CbiC in cobalamin biosynthesis
J.Struct.Biol., 153, 2006
|
|
2AFR
 
 | |
5L7H
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-methyl-5,8-dioxa-17lambda-thia-4,18-diazatetracyclo[18.2.2.1,.0]pentacosa-1(22),2(6),3,9,11,13(25),20,23-octaene-17,17-dione, Mineralocorticoid receptor, ... | | Authors: | Xue, Y, Aagaard, A, Backstrom, S, Edman, K. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
9AVK
 
 | | Structure of long Rib domain from Limosilactobacillus reuteri | | Descriptor: | SODIUM ION, THIOCYANATE ION, YSIRK signal domain/LPXTG anchor domain surface protein | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of the long Rib domain of the LPXTG-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6F3G
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-(5-propan-2-ylpyrrolo[3,2-d]pyrimidin-4-yl)cyclohexane-1,4-diamine | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3D
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3I
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | (3~{R})-3-[4-[[4-(4-ethanoylpiperazin-1-yl)cyclohexyl]amino]pyrrolo[2,1-f][1,2,4]triazin-5-yl]butanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6F3E
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 2-[(3~{R})-12-(4-morpholin-4-ylcyclohexyl)oxy-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9,11-tetraen-3-yl]ethanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
8YK7
 
 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2VB3
 
 | | Crystal structure of Ag(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, SILVER ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
2VB2
 
 | | Crystal structure of Cu(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, COPPER (II) ION, SULFATE ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
6ESN
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-acetamido-~{N}-[4-(5-cyano-3-fluoranyl-2-methoxy-phenyl)thiophen-2-yl]-2-(4-ethylsulfonylphenyl)ethanamide, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2017-10-23 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6FGQ
 
 | | Ligand complex of RORg LBD | | Descriptor: | Nuclear receptor ROR-gamma, methyl 4-[[3-[5-[2-(4-ethylsulfonylphenyl)ethanoylamino]thiophen-3-yl]pyridin-2-yl]oxymethyl]benzoate | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
1CSM
 
 | |
6RFJ
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]pyrido[3,2-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2019-04-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of a Series of 5-Azaquinazolines as Orally Efficacious IRAK4 Inhibitors Targeting MyD88L265PMutant Diffuse Large B Cell Lymphoma.
J.Med.Chem., 62, 2019
|
|
6RFI
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, methyl 4-[4-[(6-cyanoquinazolin-4-yl)amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2019-04-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Series of 5-Azaquinazolines as Orally Efficacious IRAK4 Inhibitors Targeting MyD88L265PMutant Diffuse Large B Cell Lymphoma.
J.Med.Chem., 62, 2019
|
|
6THX
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 2-[4-[(1-methylcyclopropyl)amino]-2-[(1-methylpyrazol-4-yl)amino]pyrido[3,2-d]pyrimidin-6-yl]ethanenitrile, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6TIA
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-(1-methylcyclopropyl)oxy-~{N}-[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]-6-(1-methylpyrazol-4-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6TI8
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(1-methylcyclopropyl)oxy-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[3,2-d]pyrimidine-6-carboxamide | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6THW
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 7-fluoranyl-4-(1-methylcyclopropyl)oxy-~{N}-[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6THZ
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 7-fluoranyl-~{N}-[1-(2-methyl-2-azaspiro[3.3]heptan-6-yl)pyrazol-4-yl]-4-(1-methylcyclopropyl)oxy-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Degorce, S.L. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
5NI7
 
 | | Ligand complex of RORg LBD | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, SODIUM ION, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2017-03-23 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
