5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
6IUP
 
 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
5Z1S
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(6-methoxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
5Z1T
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(6-hydroxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
6ITJ
 
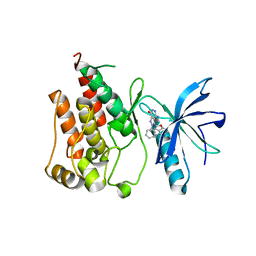 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
8HDZ
 
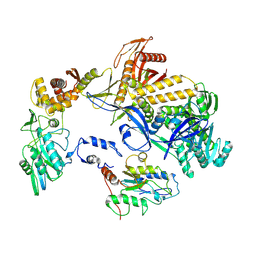 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in an apo form | | Descriptor: | A22 DNA replication processivity factor, E4 uracil-DNA glycosylase, F8 DNA polymerase | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
8HOY
 
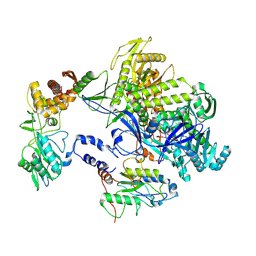 | | Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
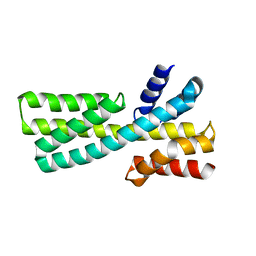 | |
7DGY
 
 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
8HPA
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
4DK0
 
 | |
2NPP
 
 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
7WLD
 
 | | Cryo-EM structure of the human glycosylphosphatidylinositol transamidase complex at 2.53 Angstrom resolution | | Descriptor: | (4S,7R)-7-[(hexadecanoyloxy)methyl]-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-3,5,8-trioxa-4lambda~5~-phosphahexacosan-1-aminium, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Luo, Y, Chao, Y, Jia, G, Zhou, Z, Su, Z, Qu, Q, Li, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins.
Nat Commun, 13, 2022
|
|
4MZD
 
 | | High resolution crystal structure of the nisin leader peptidase NisP from Lactococcus lactis | | Descriptor: | Nisin leader peptide-processing serine protease NisP | | Authors: | Rao, Z.H, Xu, Y.Y, Li, X, Yang, W. | | Deposit date: | 2013-09-30 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the nisin leader peptidase NisP revealing a C-terminal autocleavage activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3KAY
 
 | | Crystal structure of abscisic acid receptor PYL1 | | Descriptor: | Putative uncharacterized protein At5g46790 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB3
 
 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB0
 
 | | Crystal structure of abscisic acid-bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
1QCQ
 
 | | UBIQUITIN CONJUGATING ENZYME | | Descriptor: | PROTEIN (UBIQUITIN CONJUGATING ENZYME) | | Authors: | Cook, W.J, Jeffrey, L.C, Xu, Y, Chau, V. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4.
Biochemistry, 32, 1993
|
|
3KAZ
 
 | | Crystal structure of abscisic acid receptor PYL2 | | Descriptor: | 1,3-BUTANEDIOL, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
