3KO1
 
 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6DRU
 
 | | Xylosidase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glycosyl hydrolases family 31 family protein, ... | | Authors: | Cao, H, Xu, W, Betancourt, M, Walton, J.D, Brumm, P, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of alpha-Xylosidase fromAspergillus nigerin Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases.
Acs Sustain Chem Eng, 8, 2020
|
|
7N7V
 
 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
1S1G
 
 | | Crystal Structure of Kv4.3 T1 Domain | | Descriptor: | Potassium voltage-gated channel subfamily D member 3, ZINC ION | | Authors: | Scannevin, R.H, Wang, K.W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.X, Xu, Z.B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
6EF4
 
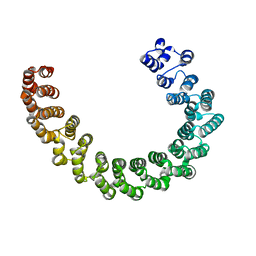 | | Crystal structure of mouse PP2A Aalpha P179R mutant | | Descriptor: | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | | Authors: | Wang, Z, Shen, G, Xu, W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Highly Recurrent PP2A A alpha-Subunit Mutation P179R Alters Protein Structure and Impairs PP2A Enzyme Function to Promote Endometrial Tumorigenesis.
Cancer Res., 79, 2019
|
|
6XN8
 
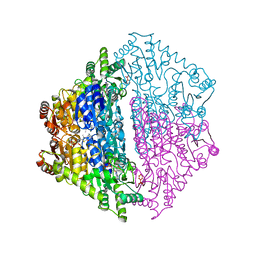 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
6XOD
 
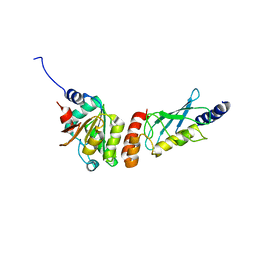 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
6PAX
 
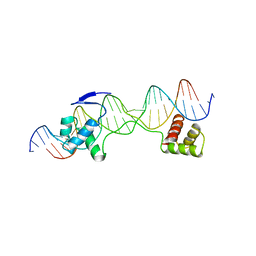 | | CRYSTAL STRUCTURE OF THE HUMAN PAX-6 PAIRED DOMAIN-DNA COMPLEX REVEALS A GENERAL MODEL FOR PAX PROTEIN-DNA INTERACTIONS | | Descriptor: | 26 NUCLEOTIDE DNA, HOMEOBOX PROTEIN PAX-6 | | Authors: | Xu, H.E, Rould, M.A, Xu, W, Epstein, J.A, Maas, R.L, Pabo, C.O. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding.
Genes Dev., 13, 1999
|
|
484D
 
 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | Descriptor: | BASIC REV PEPTIDE, RNA APTAMER | | Authors: | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
3FGA
 
 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
1LUJ
 
 | | Crystal Structure of the Beta-catenin/ICAT Complex | | Descriptor: | Beta-catenin-interacting protein 1, Catenin beta-1 | | Authors: | Graham, T.A, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2002-05-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT.
Mol.Cell, 10, 2002
|
|
1EXY
 
 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
2AZR
 
 | | Crystal structure of PTP1B with Bicyclic Thiophene inhibitor | | Descriptor: | 3-(CARBOXYMETHOXY)THIENO[2,3-B]PYRIDINE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
2B07
 
 | | Crystal structure of PTP1B with Tricyclic Thiophene inhibitor. | | Descriptor: | 6-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}-3-(CARBOXYMETHOXY)THIENO[3,2-B][1]BENZOTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
1G70
 
 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
3PUR
 
 | | CEKDM7A from C.Elegans, complex with D-2-HG | | Descriptor: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
4KK0
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
3PUQ
 
 | | CEKDM7A from C.Elegans, complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
4KK1
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
1ZXC
 
 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | Descriptor: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A8H
 
 | | Crystal structure of catalytic domain of TACE with Thiomorpholine Sulfonamide Hydroxamate inhibitor | | Descriptor: | 4-({4-[(4-AMINOBUT-2-YNYL)OXY]PHENYL}SULFONYL)-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Jin, G, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-07-08 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
