5GXH
 
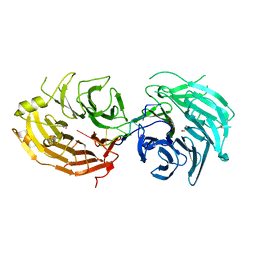 | | The structure of the Gemin5 WD40 domain with AAUUUUUG | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*G)-3'), ... | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
3UMN
 
 | | Crystal Structure of Lamin-B1 | | Descriptor: | Lamin-B1 | | Authors: | Xu, C, Bian, C.B, Amaya, M.F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-14 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1.
Febs Lett., 586, 2012
|
|
7PG9
 
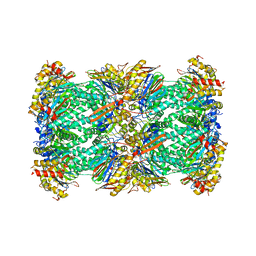 | | human 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
6TJ1
 
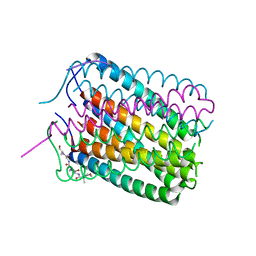 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | De novo designed WSHC6, purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-11-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
1XWN
 
 | | solution structure of cyclophilin like 1(PPIL1) and insights into its interaction with SKIP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase like 1 | | Authors: | Xu, C, Xu, Y, Tang, Y, Wu, J, Shi, Y, Huang, Q, Zhang, Q. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human peptidyl prolyl isomerase like protein 1 and insights into its interaction with SKIP
J.Biol.Chem., 281, 2006
|
|
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6P0X
 
 | |
1SJ6
 
 | | NMR Structure and Regulated Expression in APL Cell of Human SH3BGRL3 | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein 3 | | Authors: | Xu, C, Tang, Y, Xu, Y, Wu, J, Shi, Y, Zhang, Q, Zheng, P, Du, Y. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure and regulated expression in APL cell of human SH3BGRL3.
Febs Lett., 579, 2005
|
|
5VX7
 
 | |
1KJU
 
 | | Ca2+-ATPase in the E2 State | | Descriptor: | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1a | | Authors: | Xu, C, Rice, W.J, He, W, Stokes, D.L. | | Deposit date: | 2001-12-05 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A structural model for the catalytic cycle of Ca(2+)-ATPase.
J.Mol.Biol., 316, 2002
|
|
8SWI
 
 | |
8SR6
 
 | | Crystal structure of legAS4 from Legionella pneumophila subsp. pneumophila with histone H3 (3-17)peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Eukaryotic huntingtin interacting protein B, ... | | Authors: | Xu, C, Chung, I.Y.W, Cygler, M. | | Deposit date: | 2023-05-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SET and ankyrin domains of the secreted Legionella pneumophila histone methyltransferase work together to modify host chromatin.
Mbio, 14, 2023
|
|
5GXI
 
 | | Structure of the Gemin5 WD40 domain in complex with AAUUUUUGAG | | Descriptor: | Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*GP*AP*G)-3'), UNKNOWN ATOM OR ION | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
8KI4
 
 | | Crystal structure of human HSP90 in intermediate state | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein HSP 90-alpha | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
2K2W
 
 | | Second BRCT domain of NBS1 | | Descriptor: | Recombination and DNA repair protein | | Authors: | Xu, C, Cui, G, Botuyan, M, Mer, G. | | Deposit date: | 2008-04-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a second BRCT domain identified in the nijmegen breakage syndrome protein Nbs1 and its function in an MDC1-dependent localization of Nbs1 to DNA damage sites.
J.Mol.Biol., 381, 2008
|
|
3UZD
 
 | | Crystal structure of 14-3-3 GAMMA | | Descriptor: | 14-3-3 protein gamma, Histone deacetylase 4, MAGNESIUM ION, ... | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
4I79
 
 | | Crystal structure of human NUP43 | | Descriptor: | Nucleoporin Nup43, UNKNOWN ATOM OR ION, floating chain, ... | | Authors: | Xu, C, Tempel, W, Li, Z, He, H, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of human nuclear pore complex component NUP43.
Febs Lett., 589, 2015
|
|
3JZN
 
 | | Structure of EED in apo form | | Descriptor: | Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Ouyang, H, Qiu, W, MacKenzie, F, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4PXG
 
 | |
8W8K
 
 | | Crystal structures of HSP90 and the compound Ganetespid in the "closed" conformation | | Descriptor: | 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(1-methyl-1H-indol-5-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-09-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
8W4V
 
 | | Crystal structure of human HSP90 in complex with compound 4 | | Descriptor: | 4-[2-[(dimethylamino)methyl]phenyl]sulfanylbenzene-1,3-diol, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
7WEV
 
 | | SARS-COV-2 BETA VARIANT SPIKE PROTEIN IN TRANSITION STATE | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-12-24 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
4LG6
 
 | | Crystal structure of ANKRA2-CCDC8 complex | | Descriptor: | Ankyrin repeat family A protein 2, Coiled-coil domain-containing protein 8, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Bian, C, Tempel, W, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ankyrin Repeats of ANKRA2 Recognize a PxLPxL Motif on the 3M Syndrome Protein CCDC8.
Structure, 23, 2015
|
|
4LG9
 
 | | Crystal structure of TBL1XR1 WD40 repeats | | Descriptor: | F-box-like/WD repeat-containing protein TBL1XR1, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Tempel, W, He, H, Wu, X, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of TBL1XR1 WD40 repeats
TO BE PUBLISHED
|
|
7VX1
 
 | | SARS-CoV-2 Beta variant spike protein in open state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
