5SXY
 
 | |
5KSJ
 
 | | Crystal structure of deoxygenated hemoglobin in complex with Sphingosine phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
5KSI
 
 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
8GVC
 
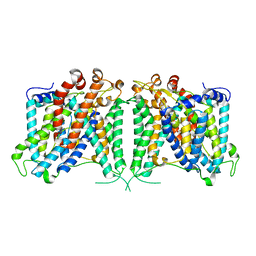 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
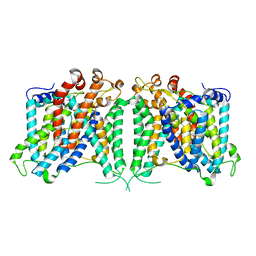 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
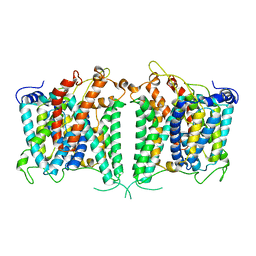 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
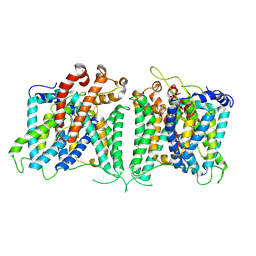 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV9
 
 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
4N1Z
 
 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1222 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Liu, Y.L, Xia, Y, Zhang, Y, Verma, I, Oldfield, E. | | Deposit date: | 2013-10-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A combination therapy for KRAS-driven lung adenocarcinomas using lipophilic bisphosphonates and rapamycin.
Sci Transl Med, 6, 2014
|
|
1IAY
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH COFACTOR PLP AND INHIBITOR AVG | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, 2-AMINO-4-(2-AMINO-ETHOXY)-BUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
7F3X
 
 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
5GNV
 
 | | Structure of PSD-95/MAP1A complex reveals unique target recognition mode of MAGUK GK domain | | Descriptor: | Disks large homolog 4, Microtubule-associated protein 1A, SULFATE ION | | Authors: | Shang, Y, Xia, Y, Zhu, R, Zhu, J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain
Biochem. J., 474, 2017
|
|
1IAX
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH PLP | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
4A4S
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*CP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4R
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GMP*GP*AP*CP*CP*CP*GP*GP*CP*UP*AP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4U
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*GP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4T
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*UP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4OR2
 
 | | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{4-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl}benzamide, CHOLESTEROL, ... | | Authors: | Wu, H, Wang, C, Gregory, K.J, Han, G.W, Cho, H.P, Xia, Y, Niswender, C.M, Katritch, V, Cherezov, V, Conn, P.J, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator
Science, 344, 2014
|
|
3WP1
 
 | | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl | | Descriptor: | Disks large homolog 4, Lethal(2) giant larvae protein homolog 2, SULFATE ION | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
3WP0
 
 | | Crystal structure of Dlg GK in complex with a phosphor-Lgl2 peptide | | Descriptor: | Disks large homolog 4, GLYCEROL, Lethal(2) giant larvae protein homolog 2 | | Authors: | Zhu, J, Shang, Y, Wan, Q, Xia, Y, Chen, J, Du, Q, Zhang, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl
Cell Res., 24, 2014
|
|
