3EZ2
 
 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
3KO1
 
 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
4FLL
 
 | | Human MetAP1 with bengamide analog YZ6, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-[(3R)-3-(furan-2-yl)-3-phenyl-propyl]-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
3S8V
 
 | | Crystal structure of LRP6-Dkk1 complex | | Descriptor: | Dickkopf-related protein 1, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3UTM
 
 | | Crystal structure of a mouse Tankyrase-Axin complex | | Descriptor: | Axin-1, Tankyrase-1 | | Authors: | Cheng, Z, Morrone, S, Xu, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8WUL
 
 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
3S94
 
 | | Crystal structure of LRP6-E1E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6CF6
 
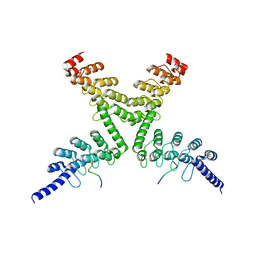 | | RNF146 TBM-Tankyrase ARC2-3 complex | | Descriptor: | RNF146, Tankyrase-1 | | Authors: | Da Rosa, P.A, Xu, W. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs.
Protein Sci., 27, 2018
|
|
3Q7J
 
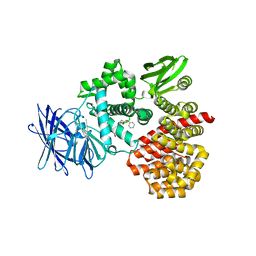 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
4N9Y
 
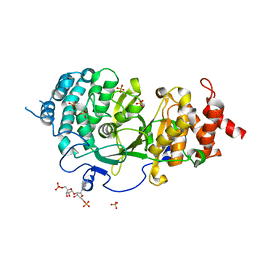 | |
4NA5
 
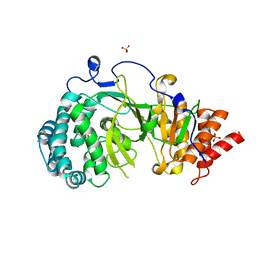 | |
3S8Z
 
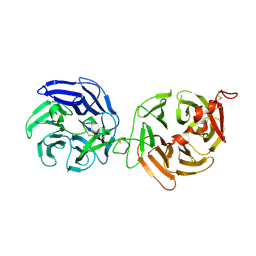 | | Crystal structure of LRP6-E3E4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4FLK
 
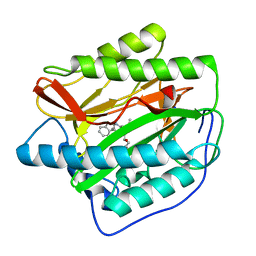 | | Human MetAP1 with bengamide analog Y10, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2,3-dihydro-1H-inden-2-yl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4FLI
 
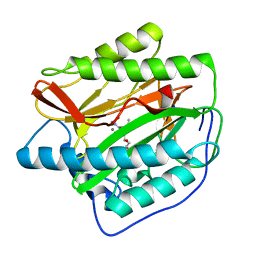 | | Human MetAP1 with bengamide analog Y16, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2-azanyl-2-oxidanylidene-ethyl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4FLJ
 
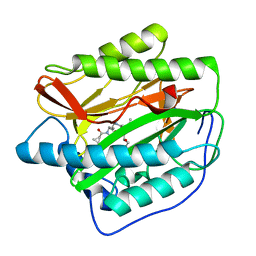 | | Human MetAP1 with bengamide analog Y08, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-[[(3S)-1-cyclopropylcarbonylpiperidin-3-yl]methyl]-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4N9Z
 
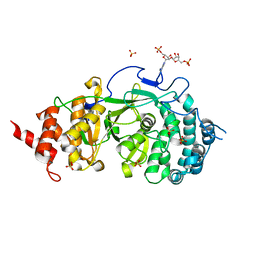 | |
3M89
 
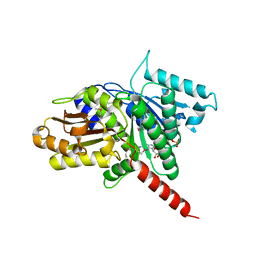 | | Structure of TubZ-GTP-g-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | Authors: | Ni, L, Xu, W, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JS6
 
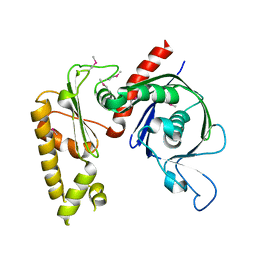 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
4EFE
 
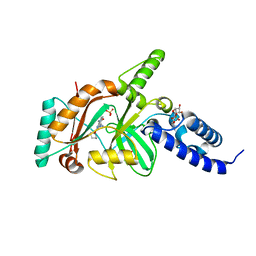 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
3MKW
 
 | | Structure of sopB(155-272)-18mer complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
4NA4
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
