1T6R
 
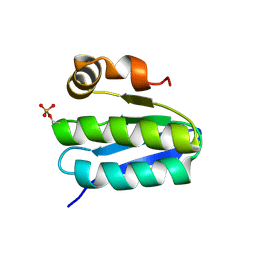 | | Solution structure of TM1442, a putative anti sigma factor antagonist in phosphorylated state | | Descriptor: | Putative anti-sigma factor antagonist TM1442 | | Authors: | Etezady-Esfarjani, T, Placzek, W, Herrmann, T, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-05-07 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the putative anti-sigma-factor antagonist TM1442 from Thermotoga maritima in the free and phosphorylated states.
Magn.Reson.Chem., 44 Spec No, 2006
|
|
1T3V
 
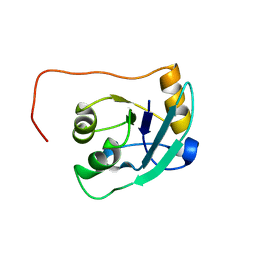 | | The NMR solution structure of TM1816 | | Descriptor: | conserved hypothetical protein | | Authors: | Columbus, L, Peti, W, Herrmann, T, Etazady, T, Klock, H, Lesley, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-04-27 | | Release date: | 2004-12-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of the conserved hypothetical protein TM1816 from Thermotoga maritima.
Proteins, 60, 2005
|
|
1U5L
 
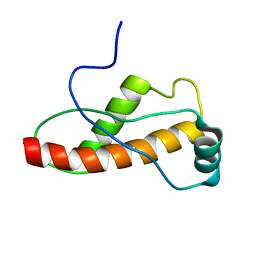 | | Solution Structure of the turtle prion protein fragment (121-226) | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chicken, turtle, and frog
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
1U3M
 
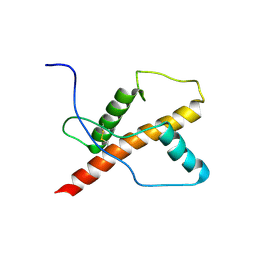 | | NMR structure of the chicken prion protein fragment 128-242 | | Descriptor: | prion-like protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-22 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chickens, turtles, and frogs
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6AQF
 
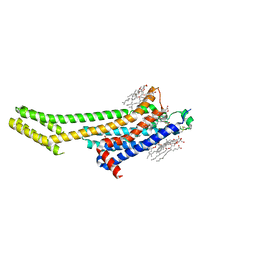 | | Crystal structure of A2AAR-BRIL in complex with the antagonist ZM241385 produced from Pichia pastoris | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Eddy, M.T, Lee, M.Y, Gao, Z, White, K, Didenko, T, Horst, R, Audet, M, Stanczak, P, McClary, K.M, Han, G.W, Jacobson, K.A, Stevens, R.C, Wuthrich, K. | | Deposit date: | 2017-08-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor.
Cell, 172, 2018
|
|
1YSY
 
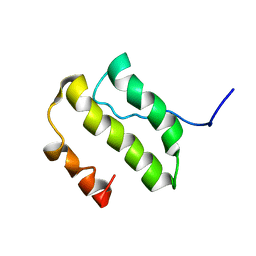 | | NMR Structure of the nonstructural Protein 7 (nsP7) from the SARS CoronaVirus | | Descriptor: | Replicase polyprotein 1ab (pp1ab) (ORF1AB) | | Authors: | Peti, W, Herrmann, T, Johnson, M.A, Kuhn, P, Stevens, R.C, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-02-09 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7.
J.Virol., 79, 2005
|
|
2MSN
 
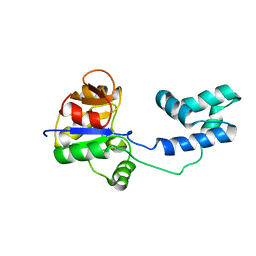 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU1
 
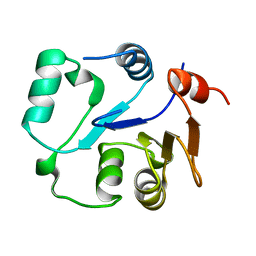 | | NMR structure of the core domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2JPO
 
 | |
2KL2
 
 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KYZ
 
 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
2KA0
 
 | | NMR structure of the protein TM1367 | | Descriptor: | uncharacterized protein TM1367 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2K9Z
 
 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KYS
 
 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
2KTS
 
 | | NMR structure of the protein NP_415897.1 | | Descriptor: | Heat shock protein hslJ | | Authors: | Serrano, P, Jaudzems, K, Geralt, M, Horst, R, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_415897.1
To be Published
|
|
2LA7
 
 | | NMR structure of the protein YP_557733.1 from Burkholderia xenovorans | | Descriptor: | Uncharacterized protein | | Authors: | Jaudzems, K, Serrano, P, Michael, G, Reto, H, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_557733.1 from Burkholderia xenovorans
To be Published
|
|
2HOA
 
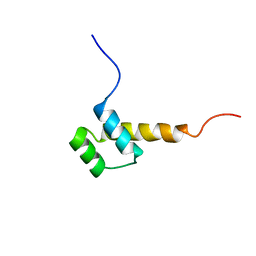 | | STRUCTURE DETERMINATION OF THE ANTP(C39->S) HOMEODOMAIN FROM NUCLEAR MAGNETIC RESONANCE DATA IN SOLUTION USING A NOVEL STRATEGY FOR THE STRUCTURE CALCULATION WITH THE PROGRAMS DIANA, CALIBA, HABAS AND GLOMSA | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Guntert, P, Qian, Y.-Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1992-04-04 | | Release date: | 1993-10-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA.
J.Mol.Biol., 217, 1991
|
|
2KQW
 
 | |
1ZE3
 
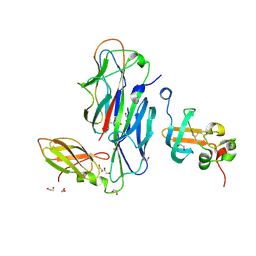 | | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, FimH protein, ... | | Authors: | Nishiyama, M, Horst, R, Eidam, O, Herrmann, T, Ignatov, O, Vetsch, M, Bettendorff, P, Jelesarov, I, Grutter, M.G, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDX
 
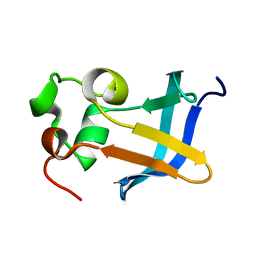 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDV
 
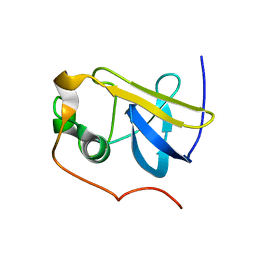 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
