7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
1ISK
 
 | | 3-OXO-DELTA5-STEROID ISOMERASE, NMR, 20 STRUCTURES | | Descriptor: | 3-OXO-DELTA5-STEROID ISOMERASE | | Authors: | Wu, Z.R, Ebrahimian, S, Zawrotny, M.E, Thornburg, L.D, Perez-Alvarado, G.C, Brothers, P, Pollack, R.M, Summers, M.F. | | Deposit date: | 1997-03-12 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 3-oxo-delta5-steroid isomerase.
Science, 276, 1997
|
|
1NAJ
 
 | | High resolution NMR Structure Of DNA Dodecamer Determined In Aqueous Dilute Liquid Crystalline Phase | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Wu, Z, Delaglio, F, Tjandra, N, Zhurkin, V, Bax, A. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and (31)P chemical shift anisotropy.
J.Biomol.Nmr, 26, 2003
|
|
1ZWO
 
 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
1ZWM
 
 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
8XGO
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGU
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGS
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
3KM4
 
 | | Optimization of Orally Bioavailable Alkyl Amine Renin Inhibitors | | Descriptor: | (3R)-3-[(1S)-4-(acetylamino)-1-(3-chlorophenyl)-1-hydroxybutyl]-N-{(1S)-2-cyclohexyl-1-[(methylamino)methyl]ethyl}piperidine-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of orally bioavailable alkyl amine renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7WJU
 
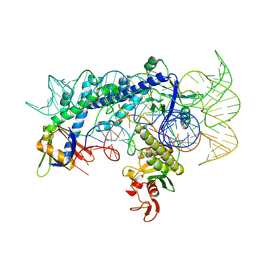 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
3K2S
 
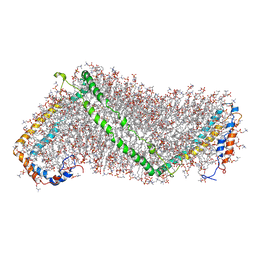 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
3GW5
 
 | | Crystal structure of human renin complexed with a novel inhibitor | | Descriptor: | (3R)-3-[(1S)-1-(3-chlorophenyl)-1-hydroxy-5-methoxypentyl]-N-{(1S)-2-cyclohexyl-1-[(methylamino)methyl]ethyl}piperidine-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2009-03-31 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and optimization of renin inhibitors: Orally bioavailable alkyl amines.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2NRF
 
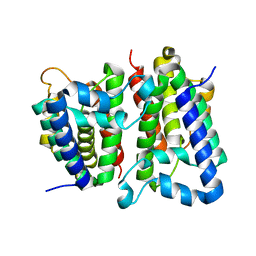 | | Crystal Structure of GlpG, a Rhomboid family intramembrane protease | | Descriptor: | Protein GlpG | | Authors: | Wu, Z, Yan, N, Feng, L, Yan, H, Gu, L, Shi, Y. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3Q3T
 
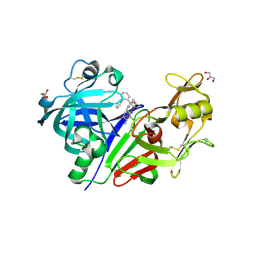 | | Alkyl Amine Renin Inhibitors: Filling S1 from S3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Wu, Z, McKeever, B. | | Deposit date: | 2010-12-22 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biphenyl/diphenyl ether renin inhibitors: Filling the S1 pocket of renin via the S3 pocket.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q4B
 
 | | Clinically Useful Alkyl Amine Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Renin, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of VTP-27999, an Alkyl Amine Renin Inhibitor with Potential for Clinical Utility.
ACS Med Chem Lett, 2, 2011
|
|
3Q5H
 
 | | Clinically Useful Alkyl Amine Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Renin, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2010-12-28 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of VTP-27999, an Alkyl Amine Renin Inhibitor with Potential for Clinical Utility.
ACS Med Chem Lett, 2, 2011
|
|
7FEF
 
 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
1QBQ
 
 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
6LBU
 
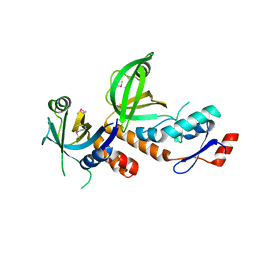 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1ZXT
 
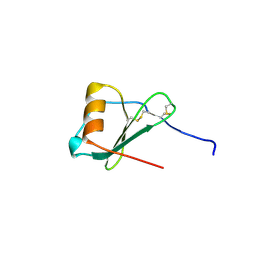 | | Crystal Structure of A Viral Chemokine | | Descriptor: | functional macrophage inflammatory protein 1-alpha homolog | | Authors: | Luz, J.G, Yu, M, Su, Y, Wu, Z, Zhou, Z, Sun, R, Wilson, I.A. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of viral macrophage inflammatory protein I encoded by Kaposi's sarcoma-associated herpesvirus at 1.7A.
J.Mol.Biol., 352, 2005
|
|
8Y3M
 
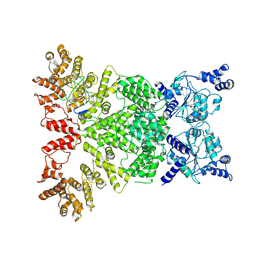 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3W
 
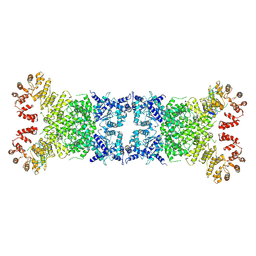 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y13
 
 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | Descriptor: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
