8HKH
 
 | |
7RNN
 
 | | Human ASIC1a-Nb.C1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Nanobodies Nb.C1 | | Authors: | Wu, Y, Chen, Z, Sigworth, F.J, Canessa, C.M. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure and analysis of nanobody binding to the human ASIC1a ion channel.
Elife, 10, 2021
|
|
8VC6
 
 | |
8VC4
 
 | |
8VC3
 
 | | Voltage gated potassium ion channel Kv1.2 in complex with DTx | | Descriptor: | Kunitz-type serine protease inhibitor homolog alpha-dendrotoxin, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2 | | Authors: | Wu, Y, Sigworth, F.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Kv1.2 potassium channels, conducting and non-conducting.
Biorxiv, 2024
|
|
8VCH
 
 | |
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
4RZV
 
 | | Crystal structure of the BRAF (R509H) kinase domain monomer bound to Vemurafenib | | Descriptor: | N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
4RZW
 
 | | Crystal structure of BRAF (R509H) kinase domain bound to AZ628 | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
1OP1
 
 | | Solution NMR structure of domain 1 of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.-X. | | Deposit date: | 2003-03-04 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
1PGC
 
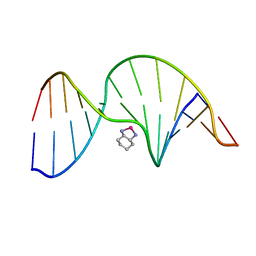 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
2MRI
 
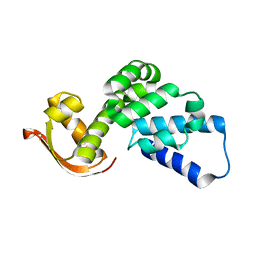 | |
2MR3
 
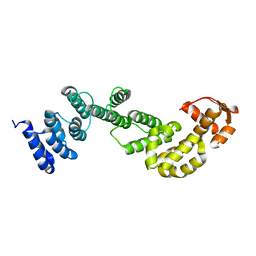 | | A subunit of 26S proteasome lid complex | | Descriptor: | 26S proteasome regulatory subunit RPN9 | | Authors: | Wu, Y, Hu, Y, Jin, C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of yeast Rpn9: insights into proteasome lid assembly.
J. Biol. Chem., 290, 2015
|
|
2MQW
 
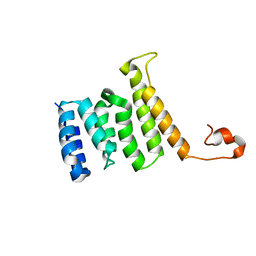 | |
5J97
 
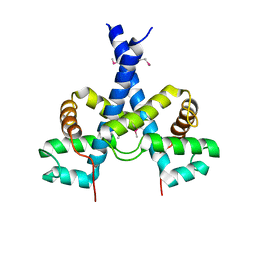 | |
5KP4
 
 | | Crystal Structure of Ketosteroid Isomerase from Pseudomonas putida (pKSI) bound to 19-nortestosterone | | Descriptor: | (8~{R},9~{S},10~{R},13~{S},14~{S},17~{S})-13-methyl-17-oxidanyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1~{H}-cyclop enta[a]phenanthren-3-one, Steroid Delta-isomerase | | Authors: | Wu, Y, Boxer, S.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | A Critical Test of the Electrostatic Contribution to Catalysis with Noncanonical Amino Acids in Ketosteroid Isomerase.
J.Am.Chem.Soc., 138, 2016
|
|
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
1OV2
 
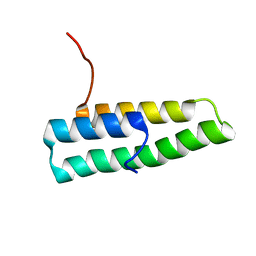 | | Ensemble of the solution structures of domain one of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
4R8W
 
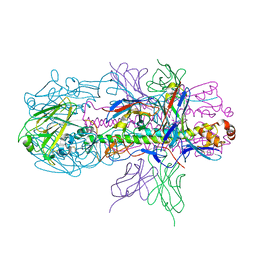 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
1PG9
 
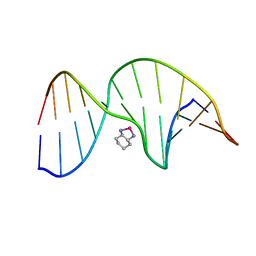 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
6UFS
 
 | |
2KQ1
 
 | | Solution Structure Of Protein BH0266 From Bacillus halodurans. Northeast Structural Genomics Consortium Target BhR97a | | Descriptor: | BH0266 protein | | Authors: | Wu, Y, Eletsky, A, Lee, D, Sathyamoorthy, B, Buchwald, W, Hua, J, Ciccosanti, C, He, Y, Hamilton, K, Acton, T, Xiao, R, Everett, J, Lee, H, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein BH0266 From Bacillus halodurans
To be Published
|
|
1XAO
 
 | | Hsp40-Ydj1 dimerization domain | | Descriptor: | Mitochondrial protein import protein MAS5 | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2004-08-26 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40
J.Mol.Biol., 346, 2005
|
|
2AS5
 
 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | Descriptor: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | Authors: | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
