7RXD
 
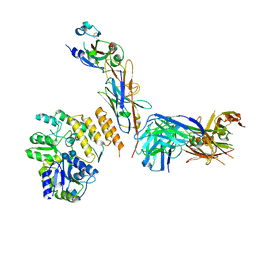 | | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | | Descriptor: | Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RXC
 
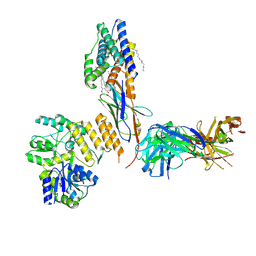 | | CryoEM structure of KDELR with Legobody | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ER lumen protein-retaining receptor 2, Fab_8D3_2 heavy chain, ... | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DSJ
 
 | |
7DSM
 
 | |
7DSO
 
 | |
7DSR
 
 | |
7DSP
 
 | |
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
7R9D
 
 | | Crystal structure of Nb_0 in complex with Fab_8D3 | | Descriptor: | Fab 8D3 heavy chain, Fab 8D3 light chain, Nanobody N0 | | Authors: | Wu, X.D, Rapoport, T.A. | | Deposit date: | 2021-06-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4A4Q
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | PROTEASE, methyl [(2S)-1-{2-(2-{(3R,4S)-3-benzyl-4-hydroxy-1-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-2-oxopyrrolidin-3-yl}ethyl)-2-[4-(pyridin-4-yl)benzyl]hydrazinyl}-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B, Unge, J, Larhed, M. | | Deposit date: | 2011-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
4A6C
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | METHYL ((S)-1-(2-(3-((3S,4S)-3-BENZYL-4-HYDROXY-1-((1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL)-2-OXOPYRROLIDIN-3-YL)PROPYL)-2-(4-(PYRIDIN-4-YL)BENZYL)HYDRAZINYL)-3,3-DIMETHYL-1-OXOBUTAN-2-YL)CARBAMATE, POL PROTEIN | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Unge, J, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B.B, Unge, J, Larhed, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
4A6B
 
 | | Stereoselective Synthesis, X-ray Analysis, and Biological Evaluation of a New Class of Lactam Based HIV-1 Protease Inhibitors | | Descriptor: | METHYL ((S)-1-(2-([1,1'-BIPHENYL]-4-YLMETHYL)-2-(3-((3S,4S)-3-BENZYL-4-HYDROXY-1-((1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL)-2-OXOPYRROLIDIN-3-YL)PROPYL)HYDRAZINYL)-3,3-DIMETHYL-1-OXOBUTAN-2-YL)CARBAMATE, POL PROTEIN | | Authors: | Wu, X, Ohrngren, P, Joshi, A.A, Trejos, A, Persson, M, Unge, J, Arvela, R.K, Wallberg, H, Vrang, L, Rosenquist, A, Samuelsson, B.B, Unge, J, Larhed, M. | | Deposit date: | 2011-11-01 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, X-Ray Analysis, and Biological Evaluation of a New Class of Stereopure Lactam-Based HIV-1 Protease Inhibitors.
J.Med.Chem., 55, 2012
|
|
1CKB
 
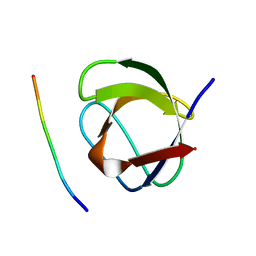 | |
1CKA
 
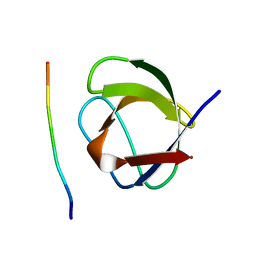 | |
8WA1
 
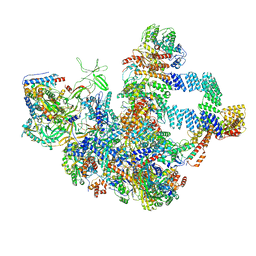 | |
8W9Z
 
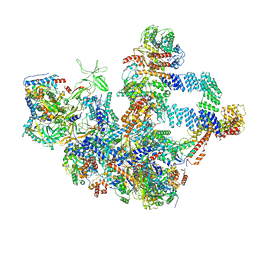 | | The cryo-EM structure of the Nicotiana tabacum PEP-PAP | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta'', ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8WA0
 
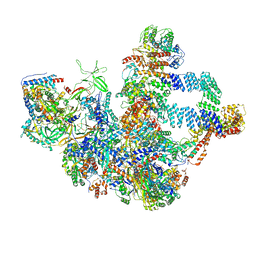 | |
8VEC
 
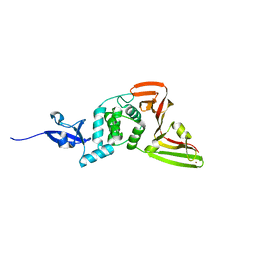 | | Deep Mutational Scanning of SARS-CoV-2 PLpro | | Descriptor: | Papain-like protease nsp3, ZINC ION | | Authors: | Wu, X, Nguyen, J.V, Call, M.E, Call, M.J. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational Profiling of SARS-CoV-2 PLpro in human cells reveals requirements for function, structure, and drug escape
Biorxiv, 2024
|
|
2C2N
 
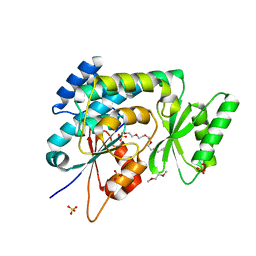 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
6ND1
 
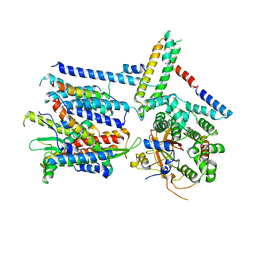 | | CryoEM structure of the Sec Complex from yeast | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Wu, X, Cabanos, C, Rapoport, T.A. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the post-translational protein translocation machinery of the ER membrane.
Nature, 566, 2019
|
|
5DSE
 
 | | Crystal Structure of the TTC7B/Hyccin Complex | | Descriptor: | Hyccin, Tetratricopeptide repeat protein 7B | | Authors: | Wu, X, Baskin, J.M, Reinisch, K.M, De Camilli, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane.
Nat.Cell Biol., 18, 2016
|
|
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | Deposit date: | 2019-03-26 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
6JHO
 
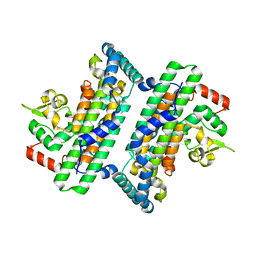 | | The complex crystal structure of Cagbeta with CagZ revealed a novel regulatory mechanism for T4SS coupling ATPase in Helicobacter pylori | | Descriptor: | Cag pathogenicity island protein (Cag5), Cag pathogenicity island protein (Cag6) | | Authors: | Wu, X, Zhao, Y, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Wu, Y. | | Deposit date: | 2019-02-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The complex crystal structure of Cagbeta with CagZ revealed a novel regulatory mechanism in VirD4 coupling ATPase
To Be Published
|
|
6VK3
 
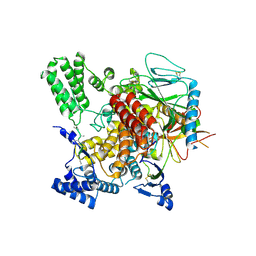 | | CryoEM structure of Hrd3/Yos9 complex | | Descriptor: | Hrd3, Protein OS-9 homolog | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
