6ZDM
 
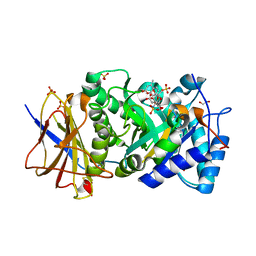 | | Crystal structure of human heparanase in complex with a N',6O'-bis-sulfated 4-methylumbelliferyl heparan sulfate disaccharide | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{S})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-4,5-bis(oxidanyl)-3-(sulfoamino)-6-(sulfooxymethyl)oxan-2-yl]oxy-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structural insights into heparanase activity using a fluorogenic heparan sulfate disaccharide.
Chem.Commun.(Camb.), 56, 2020
|
|
5LA7
 
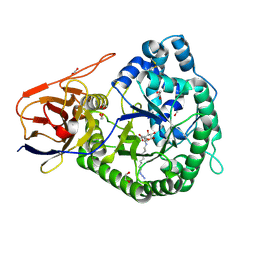 | | Crystal structure of human proheparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5L9Y
 
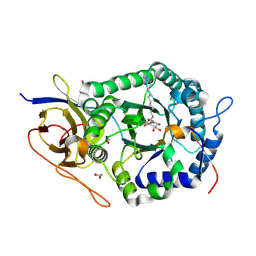 | | Crystal structure of human heparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5LA4
 
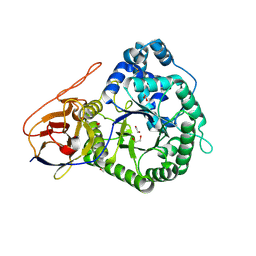 | | Crystal structure of apo human proheparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5L9Z
 
 | | Crystal structure of human heparanase nucleophile mutant (E343Q), in complex with unreacted glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
3PVL
 
 | | Structure of myosin VIIa MyTH4-FERM-SH3 in complex with the CEN1 of Sans | | Descriptor: | GLYCEROL, Myosin VIIa isoform 1, PHOSPHATE ION, ... | | Authors: | Wu, L, Pan, L.F, Wei, Z.Y, Zhang, M.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo.
Science, 331, 2011
|
|
4FTW
 
 | | Crystal structure of a carboxyl esterase N110C/L145H at 2.3 angstrom resolution | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, CHLORIDE ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4FHZ
 
 | | Crystal structure of a carboxyl esterase at 2.0 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phospholipase/Carboxylesterase, SODIUM ION | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
8HIV
 
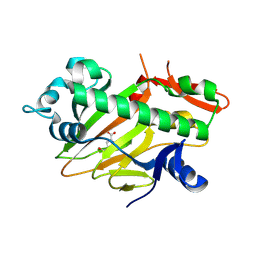 | | The structure of apo-SoBcmB with Fe(II) and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Fe/2OG dependent dioxygenase | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2022-11-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2000308 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
5CK6
 
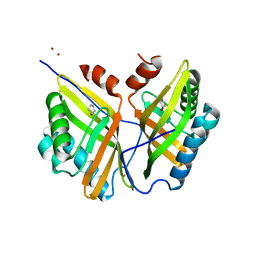 | | Crystal structure of SZ348 in complex with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, NICKEL (II) ION, ... | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2015-07-15 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SZ348 in complex with cyclopentene oxide
To Be Published
|
|
5CLK
 
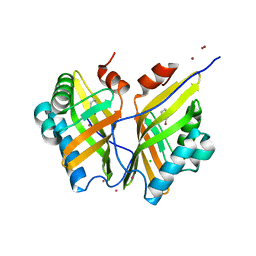 | | Crystal structure of SZ348 in complex with S,S-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CHLORIDE ION, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structure of SZ348 in complex with S,S-cyclohexanediol
To Be Published
|
|
5CF1
 
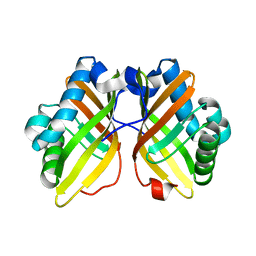 | |
5CF2
 
 | |
5E9B
 
 | | Crystal structure of human heparanase in complex with HepMer M09S05a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E98
 
 | |
5E9C
 
 | | Crystal structure of human heparanase in complex with heparin tetrasaccharide dp4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5D74
 
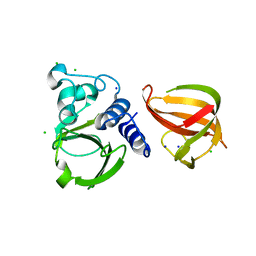 | |
5E8M
 
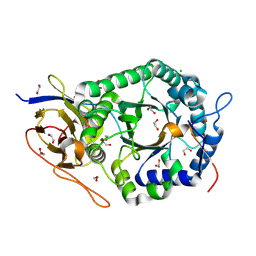 | | Crystal structure of human heparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E97
 
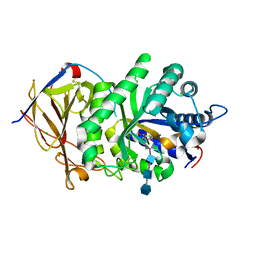 | | Glycoside Hydrolase ligand structure 1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5D76
 
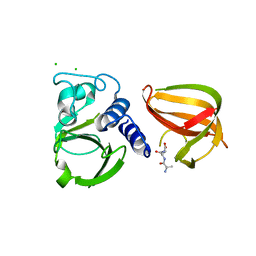 | |
5GKW
 
 | | crystal structure of SZ529 complex with (R,R)-cyclopentanediol | | Descriptor: | (1~{R},2~{R})-cyclopentane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | crystal structure of SZ529 complex with (R,R)-cyclopentanediol
To Be Published
|
|
5I23
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
5I24
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF021 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
7V2U
 
 | | The complex structure of SoBcmB and its product 2f | | Descriptor: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.00009918 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V34
 
 | | The complex structure of soBcmB and its product 1d | | Descriptor: | (3S,4S,5S,8S)-8-[(2S)-butan-2-yl]-3-methyl-3,4-bis(oxidanyl)-1-oxa-7,10-diazaspiro[4.5]decane-6,9-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.000169 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
