7E34
 
 | | Crystal structure of SUN1-Speedy A-CDK2 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, SUN domain-containing protein 1, ... | | Authors: | Chen, Y, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Nat Commun, 12, 2021
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
4KVG
 
 | | Crystal structure of RIAM RA-PH domains in complex with GTP bound Rap1 | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, H, Chang, Y.E, Brennan, M.L, Wu, J. | | Deposit date: | 2013-05-22 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism.
J Mol Cell Biol, 6, 2014
|
|
4Q2E
 
 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S258C, active mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
4JV4
 
 | | Crystal Structure of RIalpha(91-379) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
6AH3
 
 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
5WZK
 
 | | Structure of APUM23-deletion-of-insert-region-GGAAUUGACGG | | Descriptor: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | Authors: | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-01-18 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
4Q2G
 
 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S223C, inactive mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, Phosphatidate cytidylyltransferase, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
5Y1U
 
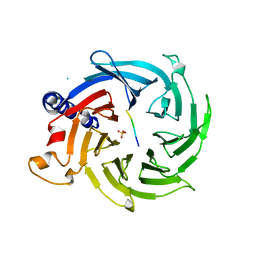 | | Crystal structure of RBBP4 bound to AEBP2 RRK motif | | Descriptor: | Histone-binding protein RBBP4, SULFATE ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2017-07-21 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4
Protein Cell, 2017
|
|
4M68
 
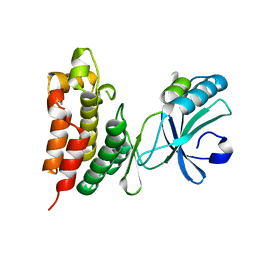 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4NTS
 
 | |
6AGB
 
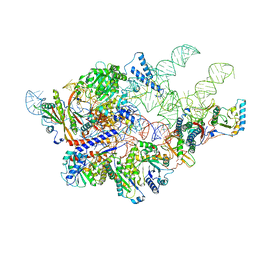 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
4NTT
 
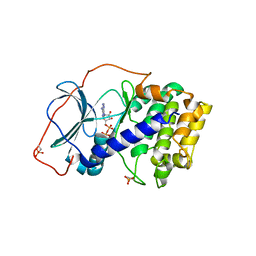 | |
4M66
 
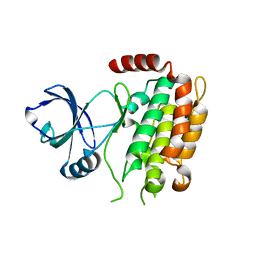 | | Crystal structure of the mouse RIP3 kinase domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4R7A
 
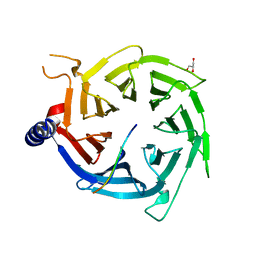 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
4M67
 
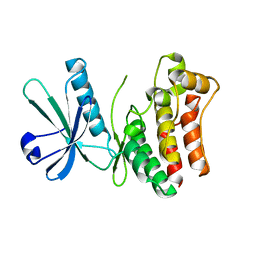 | | Crystal structure of the human MLKL kinase-like domain | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
2LW7
 
 | | NMR solution structure of human HisRS splice variant | | Descriptor: | Histidine--tRNA ligase, cytoplasmic | | Authors: | Ye, F, Wei, Z, Wu, J, Schimmel, P, Zhang, M. | | Deposit date: | 2012-07-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of human HisRS splice variant
To be Published
|
|
2K7N
 
 | |
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | Descriptor: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0Y
 
 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0X
 
 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1L
 
 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
5SYO
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, Soluble acetylcholine receptor, Neuronal acetylcholine receptor subunit alpha-3 chimera | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine
To Be Published
|
|
