1GN3
 
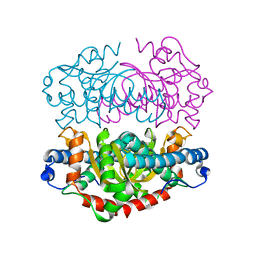 | | H145Q mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants.
Eur.J.Biochem., 251, 1998
|
|
1GJP
 
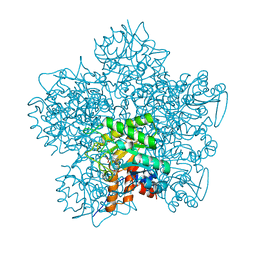 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-OXOSEBACIC ACID | | Descriptor: | 4-OXODECANEDIOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1H7N
 
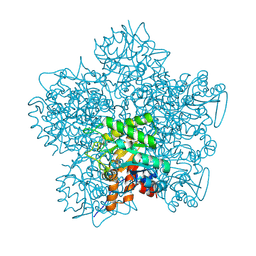 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH LAEVULINIC ACID AT 1.6 A RESOLUTION | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
1MPP
 
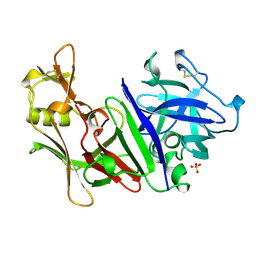 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
1N28
 
 | | Crystal structure of the H48Q mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
1MOM
 
 | | CRYSTAL STRUCTURE OF MOMORDIN, A TYPE I RIBOSOME INACTIVATING PROTEIN FROM THE SEEDS OF MOMORDICA CHARANTIA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOMORDIN | | Authors: | Husain, J, Tickle, I.J, Wood, S.P. | | Deposit date: | 1994-03-04 | | Release date: | 1994-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia.
FEBS Lett., 342, 1994
|
|
1N29
 
 | | Crystal structure of the N1A mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
5OV6
 
 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OV5
 
 | | Bacillus megaterium porphobilinogen deaminase D82E mutant | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT1
 
 | | The type III pullulan hydrolase from Thermococcus kodakarensis | | Descriptor: | CALCIUM ION, Pullulanase type II, GH13 family | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Keegan, R, Ahmad, N, Muhammad, M.A, Rashid, N, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OT0
 
 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OV4
 
 | | Bacillus megaterium porphobilinogen deaminase D82A mutant | | Descriptor: | Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1QNV
 
 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QML
 
 | | Hg complex of yeast 5-aminolaevulinic acid dehydratase | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, MERCURY (II) ION | | Authors: | Erskine, P.T, Senior, N, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-02 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3PHV
 
 | | X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES | | Descriptor: | UNLIGANDED HIV-1 PROTEASE | | Authors: | Lapatto, R, Blundell, T.L, Hemmings, A, Wilderspin, A, Wood, S.P, Danley, D.E, Geoghegan, K.F, Hawrylik, S.J, Hobart, P.M. | | Deposit date: | 1991-11-04 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes.
Nature, 342, 1989
|
|
4P9G
 
 | | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. | | Descriptor: | 2,4'-dihydroxyacetophenone dioxygenase, CARBONATE ION, FE (III) ION, ... | | Authors: | Keegan, R, Lebedev, A, Erskine, P, Guo, J, Wood, S.P, Hopper, D.J, Cooper, J.B. | | Deposit date: | 2014-04-03 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the 2,4'-dihydroxyacetophenone dioxygenase from Alcaligenes sp. 4HAP
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HTG
 
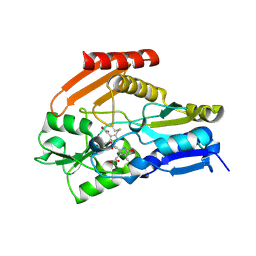 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3EQ1
 
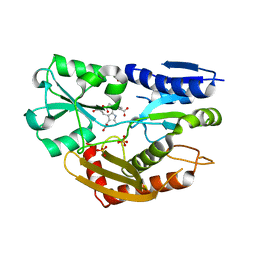 | | The Crystal Structure of Human Porphobilinogen Deaminase at 2.8A resolution | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | Authors: | Kolstoe, S.E, Gill, R, Mohammed, F, Wood, S.P. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human porphobilinogen deaminase at 2.8 A: the molecular basis of acute intermittent porphyria
Biochem.J., 420, 2009
|
|
3NFT
 
 | | Near-atomic resolution analysis of BipD- A component of the type-III secretion system of Burkholderia pseudomallei | | Descriptor: | Translocator protein bipD | | Authors: | Pal, M, Erskine, P.T, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4AVV
 
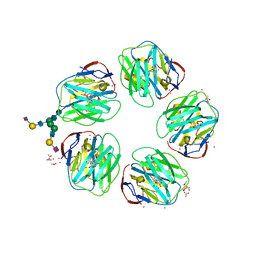 | | Structure of CPHPC bound to Serum Amyloid P Component | | Descriptor: | (2R)-1-[6-[(2R)-2-carboxypyrrolidin-1-yl]-6-oxidanylidene-hexanoyl]pyrrolidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kolstoe, S.E, Jenvey, M.C, Wood, S.P. | | Deposit date: | 2012-05-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AVS
 
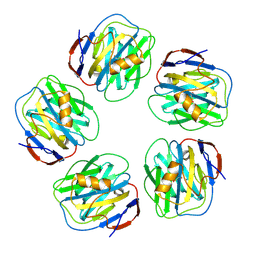 | |
4AVT
 
 | | Structure of CPHPC bound to Serum Amyloid P Component | | Descriptor: | (2R)-1-[6-[(2R)-2-carboxypyrrolidin-1-yl]-6-oxidanylidene-hexanoyl]pyrrolidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kolstoe, S.E, Purvis, A, Wood, S.P. | | Deposit date: | 2012-05-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AYU
 
 | | Structure of N-Acetyl-D-Proline bound to serum amyloid P component | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Hughes, P, Kolstoe, S.E, Wood, S.P. | | Deposit date: | 2012-06-22 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of Serum Amyloid P Component with Hexanoyl Bis(D-Proline) (Cphpc)
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LP9
 
 | | Endothiapepsin complexed with Phe-reduced-Tyr peptide. | | Descriptor: | Endothiapepsin, GLYCEROL, SULFATE ION, ... | | Authors: | Guo, J, Cooper, J.B, Wood, S.P. | | Deposit date: | 2013-07-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4MLV
 
 | | Crystal Structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
