6BVT
 
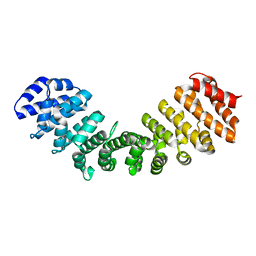 | | Importin alpha 1 in cargo free state | | Descriptor: | Importin subunit alpha-1 | | Authors: | Smith, K.M, Tsimablyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BW1
 
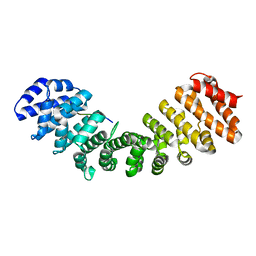 | | Hendra virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BW0
 
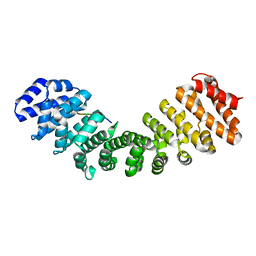 | | Nipah virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BWB
 
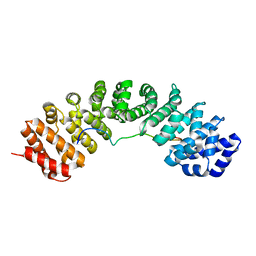 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
8U36
 
 | |
6BW9
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 1 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BVV
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
8TUV
 
 | | Merkel Cell Polyomavirus LTA NLSm bound to importin alpha 2 | | Descriptor: | 30S ribosomal protein S11, Importin subunit alpha-1, SER-ARG-LYS-ARG-LYS-PHE | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8TUU
 
 | |
8TUT
 
 | | Merkel Cell Polyomavirus LTA NLSct bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, PRO-PRO-LYS-PRO-LYS-LYS-ASN-ARG-GLU | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8TUQ
 
 | | STL Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Donnelly, C.M, Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8TUS
 
 | | WU Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Donnelly, C.M, Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-17 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8TUR
 
 | |
5SVZ
 
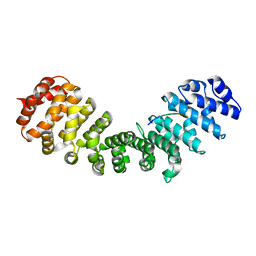 | |
4IEN
 
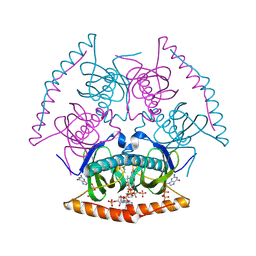 | | Crystal Structure of Acyl-CoA Hydrolase from Neisseria meningitidis FAM18 | | Descriptor: | CHLORIDE ION, COENZYME A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Khandokar, Y.B, Forwood, J.K. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, purification and crystallization of acetyl-CoA hydrolase from Neisseria meningitidis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
7KWQ
 
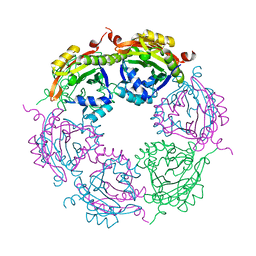 | | Spermidine N-acetyltransferase SpeG R149-K152 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KWX
 
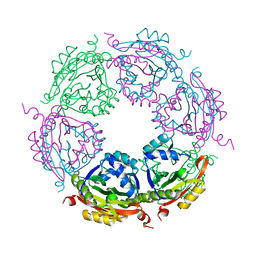 | | Spermidine N-acetyltransferase SpeG N152L mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KWJ
 
 | | Spermidine N-acetyltransferase SpeG K23-Q34 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7L04
 
 | |
7JJK
 
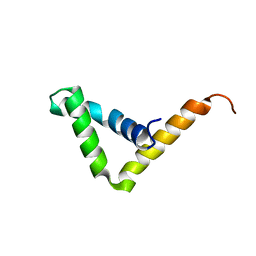 | | Crystal structure of SOX30 | | Descriptor: | Transcription factor SOX-30 | | Authors: | Ghafoori, S.M, Forwood, J.K. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of SOX30 at 1.40 Angstroms resolution
To Be Published
|
|
4QAV
 
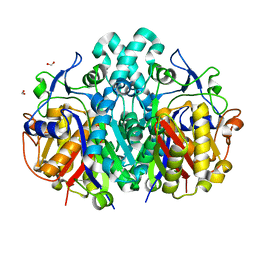 | |
2CMO
 
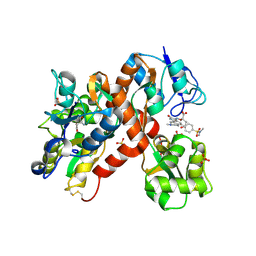 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
7KX3
 
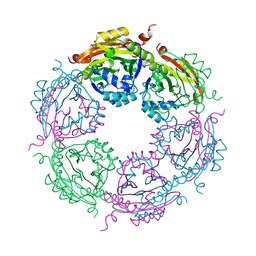 | | SpeG Spermidine N-acetyltransferase F149G mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KX2
 
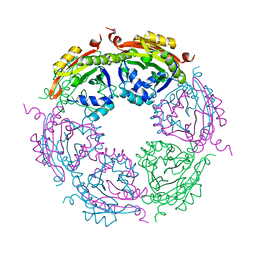 | | Spermidine N-acetyltransferase SpeG F149A mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
4M8S
 
 | |
