1B2P
 
 | | NATIVE MANNOSE-SPECIFIC BULB LECTIN FROM SCILLA CAMPANULATA (BLUEBELL) AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (LECTIN) | | Authors: | Wood, S.D, Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, Peumans, W.J, Van Damme, E.J.M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the native (unligated) mannose-specific bulb lectin from Scilla campanulata (bluebell) at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2ITO
 
 | | Crystal structure of EGFR kinase domain G719S mutation in complex with Iressa | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITU
 
 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITZ
 
 | | Crystal structure of EGFR kinase domain L858R mutation in complex with Iressa | | Descriptor: | CHLORIDE ION, EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITX
 
 | | Crystal structure of EGFR kinase domain in complex with AMP-PNP | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITY
 
 | | Crystal structure of EGFR kinase domain in complex with Iressa | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, Gefitinib | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITP
 
 | | Crystal structure of EGFR kinase domain G719S mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR PRECURSOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITW
 
 | | Crystal structure of EGFR kinase domain in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITT
 
 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITQ
 
 | | Crystal structure of EGFR kinase domain G719S mutation in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITV
 
 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AMP-PNP | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITN
 
 | | Crystal structure of EGFR kinase domain G719S mutation in complex with AMP-PNP | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2J6M
 
 | | Crystal structure of EGFR kinase domain in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-09-29 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
1QNV
 
 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QML
 
 | | Hg complex of yeast 5-aminolaevulinic acid dehydratase | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, MERCURY (II) ION | | Authors: | Erskine, P.T, Senior, N, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-02 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1GYK
 
 | | Serum Amyloid P Component co-crystallised with MOBDG at neutral pH | | Descriptor: | CALCIUM ION, METHYL 4,6-O-[(1R)-1-CARBOXYETHYLIDENE]-BETA-D-GALACTOPYRANOSIDE, SERUM AMYLOID P-COMPONENT | | Authors: | Thompson, D, Pepys, M.B, Tickle, I, Wood, S.P. | | Deposit date: | 2002-04-25 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of Crystalline Complexes of Human Serum Amyloid P Component with its Carbohydrate Ligand, the Cyclic Pyruvate Acetal of Galactose
J.Mol.Biol., 320, 2002
|
|
4LP9
 
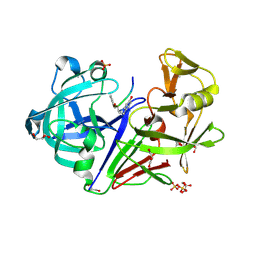 | | Endothiapepsin complexed with Phe-reduced-Tyr peptide. | | Descriptor: | Endothiapepsin, GLYCEROL, SULFATE ION, ... | | Authors: | Guo, J, Cooper, J.B, Wood, S.P. | | Deposit date: | 2013-07-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of endothiapepsin complexed with a Phe-Tyr reduced-bond inhibitor at 1.35 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4MLV
 
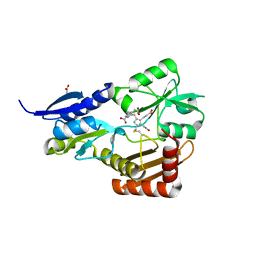 | | Crystal Structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MLQ
 
 | | Crystal structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3KQR
 
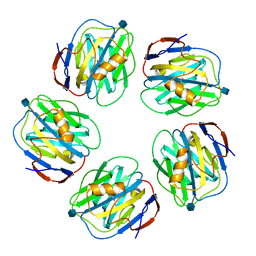 | | The structure of serum amyloid p component bound to phosphoethanolamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-11-17 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
1H7R
 
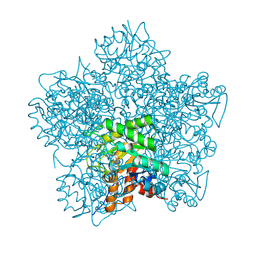 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH SUCCINYLACETONE AT 2.0 A RESOLUTION. | | Descriptor: | 4,6-DIOXOHEPTANOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Newbold, R, Brindley, A.A, Wood, S.P, Shoolingin-Jordan, P.M, Warren, M.J, Cooper, J.B. | | Deposit date: | 2001-07-09 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Substrate and Three Inhibitors
J.Mol.Biol., 312, 2001
|
|
5OV5
 
 | | Bacillus megaterium porphobilinogen deaminase D82E mutant | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT1
 
 | | The type III pullulan hydrolase from Thermococcus kodakarensis | | Descriptor: | CALCIUM ION, Pullulanase type II, GH13 family | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Keegan, R, Ahmad, N, Muhammad, M.A, Rashid, N, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OV6
 
 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OV4
 
 | | Bacillus megaterium porphobilinogen deaminase D82A mutant | | Descriptor: | Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
