5J9F
 
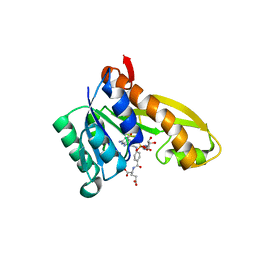 | | Human GAR transformylase in complex with GAR and (4-{[2-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzoyl)-L-glutamic acid (AGF183) | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-(4-{[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzene-1-carbonyl)-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong, J, Deis, S.M, Dann III, C.E. | | Deposit date: | 2016-04-09 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
J.Med.Chem., 59, 2016
|
|
4XCM
 
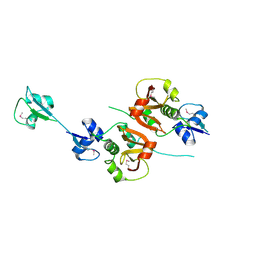 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YH2
 
 | |
4YJY
 
 | |
4S11
 
 | |
4S10
 
 | |
6IM8
 
 | | CueO-PM2 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM9
 
 | | MDM2 bound CueO-PM2 sensor | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM7
 
 | | CueO-12.1 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
3F6F
 
 | |
3F63
 
 | |
3F6D
 
 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, F123A, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3G7I
 
 | |
3G7J
 
 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, Y119E, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3GH6
 
 | |
5WTS
 
 | |
3MAK
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
4QPQ
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | Relaxosome protein TraM, sbmA DNA1, sbmA DNA2 | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4QPO
 
 | | Mechanistic basis of plasmid-specific DNA binding of the F plasmid regulatory protein, TraM | | Descriptor: | PHOSPHATE ION, Relaxosome protein TraM | | Authors: | Peng, Y, Lu, J, Wong, J, Edwards, R.A, Frost, L.S, Glover, J.N.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanistic Basis of Plasmid-Specific DNA Binding of the F Plasmid Regulatory Protein, TraM.
J.Mol.Biol., 426, 2014
|
|
4PW6
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW5
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW7
 
 | | structure of UHRF2-SRA in complex with a 5mC-containing DNA | | Descriptor: | 5mC-containing DNA1, 5mC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
8QVS
 
 | | Crystal structure of Ompk36 GD at 3500 eV based on a combination of spherical harmonics and analytical absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUR
 
 | | Crystal structure of Ompk36 GD at 3500 eV with no absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
