1SN7
 
 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
9CBE
 
 | |
2ION
 
 | |
2IOL
 
 | |
1ZTH
 
 | |
1ZTF
 
 | |
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6DWF
 
 | |
8HVP
 
 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
1N00
 
 | | Annexin Gh1 from cotton | | Descriptor: | SULFATE ION, annexin Gh1 | | Authors: | Hofmann, A, Delmer, D.P, Wlodawer, A. | | Deposit date: | 2002-10-10 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of annexin Gh1 from Gossypium hirsutum reveals an unusual S3 cluster.
Eur.J.Biochem., 270, 2003
|
|
1N1F
 
 | | Crystal Structure of Human Interleukin-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, interleukin-19 | | Authors: | Chang, C, Magracheva, E, Kozlov, S, Fong, S, Tobin, G, Kotenko, S, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of interleukin-19 defines a new subfamily of helical cytokines
J.Biol.Chem., 278, 2003
|
|
7HVP
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1EMM
 
 | |
1EML
 
 | |
1EMC
 
 | |
6RIZ
 
 | | Crystal structure of gp41-1 intein (C1A, F65W, D107C) | | Descriptor: | gp41-1 intein | | Authors: | Beyer, H.M, Lountos, G.T, Mikula, M.K, Wlodawer, A, Iwai, H. | | Deposit date: | 2019-04-25 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms.
Int J Mol Sci, 21, 2020
|
|
2GTY
 
 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
4RLD
 
 | |
6FIV
 
 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
7UT3
 
 | | Crystal structure of complex of Fab, G10C with GalNAc-pNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside, Fab protein heavy chain, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Development of a GalNAc-Tyrosine-Specific Monoclonal Antibody and Detection of Tyrosine O -GalNAcylation in Numerous Human Tissues and Cell Lines.
J.Am.Chem.Soc., 144, 2022
|
|
8RSA
 
 | |
7UPZ
 
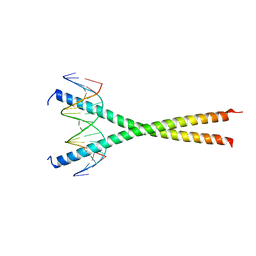 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
5L04
 
 | |
