5T5O
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
1B11
 
 | | STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH TL-3-093 | | Descriptor: | PROTEIN (Feline Immunodeficiency Virus PROTEASE), SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Gustchina, A, Li, M, Wlodawer, A. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
3QVC
 
 | |
1HO3
 
 | | CRYSTAL STRUCTURE ANALYSIS OF E. COLI L-ASPARAGINASE II (Y25F MUTANT) | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE II | | Authors: | Jaskolski, M, Kozak, M, Lubkowski, P, Palm, J.G, Wlodawer, A. | | Deposit date: | 2000-12-08 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two highly homologous bacterial L-asparaginases: a case of enantiomorphic space groups.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3GEF
 
 | | Crystal structure of the R482W mutant of lamin A/C | | Descriptor: | Lamin-A/C | | Authors: | Magracheva, E, Kozlov, S, Stuart, C, Wlodawer, A, Zdanov, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the lamin A/C R482W mutant responsible for dominant familial partial lipodystrophy (FPLD).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5VF2
 
 | | scFv 2D10 re-refined as a complex with trehalose replacing the original alpha-1,6-mannobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
3SM1
 
 | |
2GTY
 
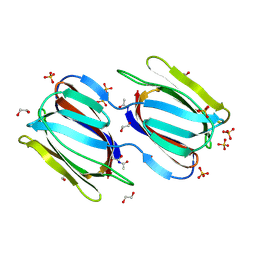 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GUD
 
 | |
2HYR
 
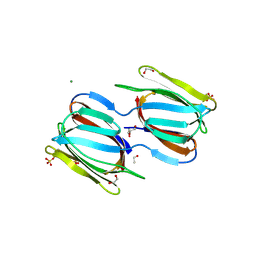 | | Crystal structure of a complex of griffithsin with maltose | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, MAGNESIUM ION, ... | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-08-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin.
Proteins, 67, 2007
|
|
3QS1
 
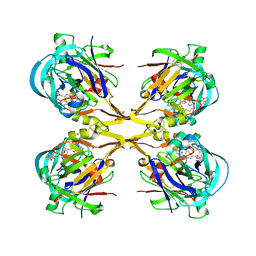 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
3EZM
 
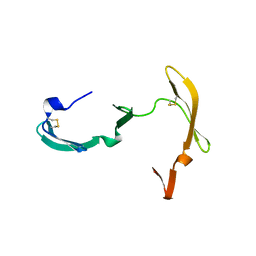 | | CYANOVIRIN-N | | Descriptor: | PROTEIN (CYANOVIRIN-N) | | Authors: | Yang, F, Wlodawer, A. | | Deposit date: | 1998-12-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping.
J.Mol.Biol., 288, 1999
|
|
3FNS
 
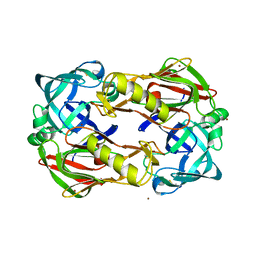 | |
3FNT
 
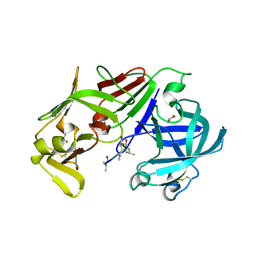 | | Crystal structure of pepstatin A bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, HAP protein, Inhibitor, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
3RFI
 
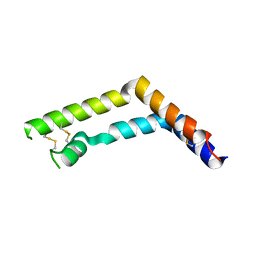 | |
3SLZ
 
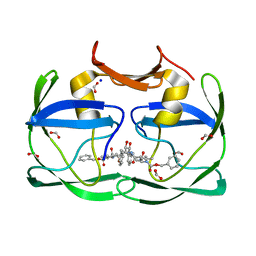 | | The crystal structure of XMRV protease complexed with TL-3 | | Descriptor: | FORMIC ACID, SODIUM ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3SM2
 
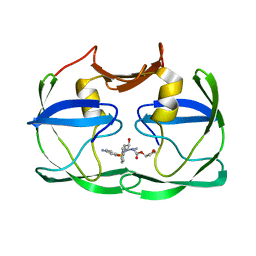 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
2GUC
 
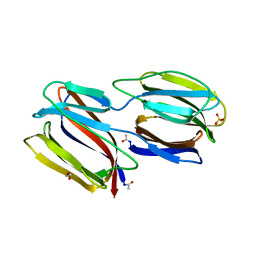 | |
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HEX
 
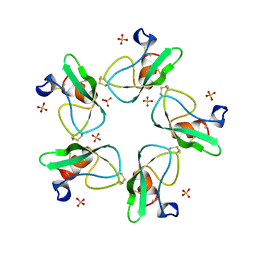 | |
2GUE
 
 | |
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3OG6
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
