8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SDF
 
 | |
8SDG
 
 | |
3QG7
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Antibody Heavy Chain, AP4-24H11 Antibody Light Chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Kirchdoerfer, R.K, Kaufmann, G.F, Janda, J.D, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
8SD4
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 7 | | Descriptor: | (S~1~S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-[(dimethylamino)methyl]azetidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Kadam, R.U, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SD2
 
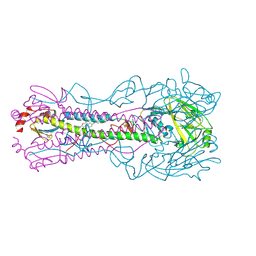 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 4 | | Descriptor: | (S~6~S)-N-{3-chloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-5,7-dihydro-6H-pyrrolo[3,4-b]pyridine-6-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadam, R.U, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3QL3
 
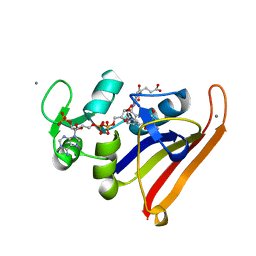 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
8SDH
 
 | |
3QG6
 
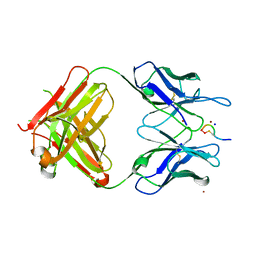 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Heavy Chain, AP4-24H11 Light Chain, Agr autoinducing peptide, ... | | Authors: | Kirchdoerfer, R.N, Janda, J.D, Kaufmann, G.F, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
8EQA
 
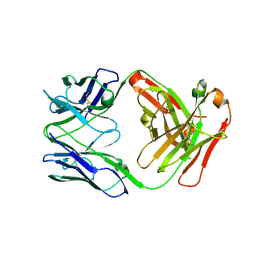 | | Crystal structure of human anti-N1 neuraminidase 2H08 Fab | | Descriptor: | 2H08 Fab heavy chain, 2H08 Fab light chain, ZINC ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8EQC
 
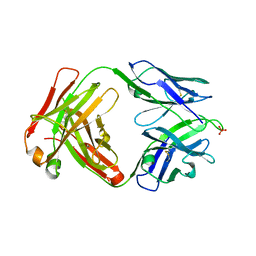 | | Crystal structure of human anti-N1 neuraminidase 3H03 Fab | | Descriptor: | 3H03 Fab heavy chain, 3H03 Fab light chain, PHOSPHATE ION | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8TJ4
 
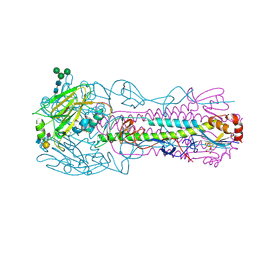 | | CRYSTAL STRUCTURE OF THE A/Bangkok/1/1979(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
3Q1S
 
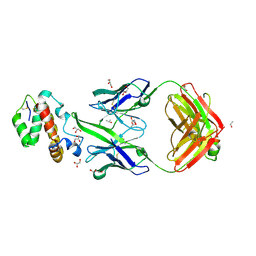 | | HIV-1 neutralizing antibody Z13e1 in complex with epitope display protein | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Interleukin-22, ... | | Authors: | Stanfield, R.L, Julien, J.-P, Pejchal, R, Gach, J.S, Zwick, M.B, Wilson, I.A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of a Protein Immunogen that Displays an HIV-1 gp41 Neutralizing Epitope.
J.Mol.Biol., 414, 2011
|
|
8TJ8
 
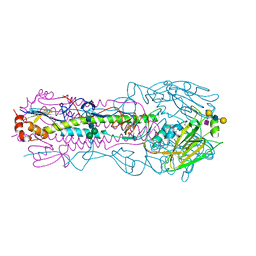 | | CRYSTAL STRUCTURE OF THE A/Moscow/10/1999(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ9
 
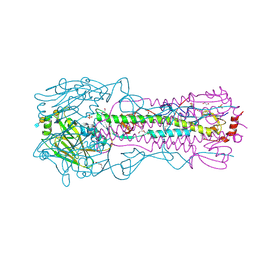 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ7
 
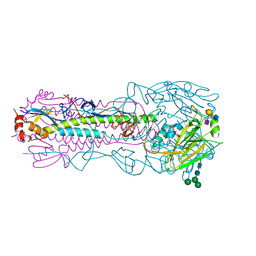 | | CRYSTAL STRUCTURE OF THE A/Shandong/9/1993(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJB
 
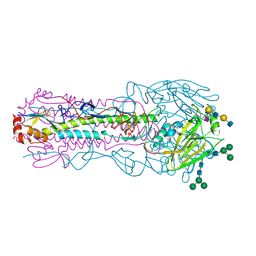 | | CRYSTAL STRUCTURE OF THE A/Texas/73/2017(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ6
 
 | | CRYSTAL STRUCTURE OF THE A/Beijing/353/1989(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJA
 
 | | CRYSTAL STRUCTURE OF THE A/Ecuador/1374/2016(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
3OZ9
 
 | |
8GB6
 
 | |
8GB7
 
 | |
8GB8
 
 | |
8GB5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | Descriptor: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-02-24 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
