5FBZ
 
 | | Structure of subtilase SubHal from Bacillus halmapalus - complex with chymotrypsin inhibitor CI2A | | Descriptor: | Autoproteolytic fragment of enzyme subtilase SubHal, CALCIUM ION, Enzyme subtilase SubHal from Bacillus halmapalus, ... | | Authors: | Dohnalek, J, Brzozowski, A.M, Svendsen, A, Wilson, K.S. | | Deposit date: | 2015-12-14 | | Release date: | 2016-05-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
5FJJ
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FFN
 
 | | Complex of subtilase SubTY from Bacillus sp. TY145 with chymotrypsin inhibitor CI2A | | Descriptor: | CALCIUM ION, Enzyme subtilase SubTY from Bacillus sp. TY145, SODIUM ION, ... | | Authors: | McAuley, K.E, Svendsen, A, Oestergaard, P.R, Dohnalek, J, Wilson, K.S. | | Deposit date: | 2015-12-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
4LZT
 
 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2TPL
 
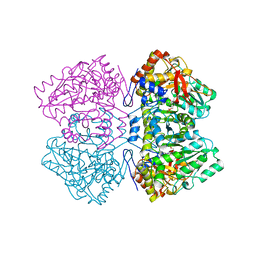 | | TYROSINE PHENOL-LYASE FROM CITROBACTER INTERMEDIUS COMPLEX WITH 3-(4'-HYDROXYPHENYL)PROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE AND CS+ ION | | Descriptor: | CESIUM ION, HYDROXYPHENYL PROPIONIC ACID, TYROSINE PHENOL-LYASE | | Authors: | Antson, A.A, Demidkina, T.V, Wilson, K.S. | | Deposit date: | 1997-01-23 | | Release date: | 1997-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Citrobacter freundii tyrosine phenol-lyase complexed with 3-(4'-hydroxyphenyl)propionic acid, together with site-directed mutagenesis and kinetic analysis, demonstrates that arginine 381 is required for substrate specificity.
Biochemistry, 36, 1997
|
|
2TEC
 
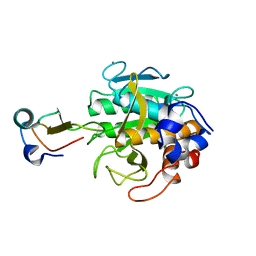 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2VK2
 
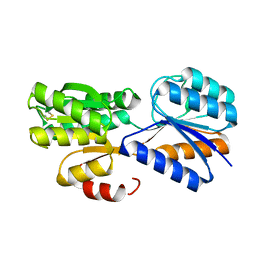 | | Crystal structure of a galactofuranose binding protein | | Descriptor: | ABC TRANSPORTER PERIPLASMIC-BINDING PROTEIN YTFQ, beta-D-galactofuranose | | Authors: | Muller, A, Horler, R.S.P, Thomas, G.H, Wilson, K.S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Furanose-Specific Sugar Transport: Characterization of a Bacterial Galactofuranose-Binding Protein.
J.Biol.Chem., 284, 2009
|
|
2RVE
 
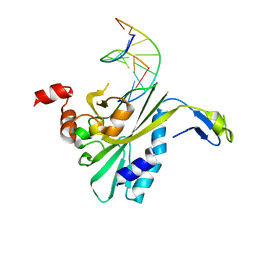 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*CP*TP*CP*G)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilson, K.S. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
2PFM
 
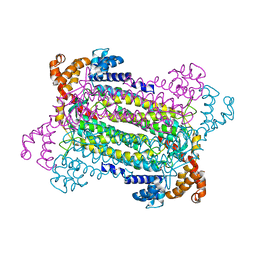 | | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis | | Descriptor: | Adenylosuccinate lyase, MALONATE ION | | Authors: | Levdikov, V.M, Blagova, E.V, Baumgart, M, Moroz, O.V, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis
To be Published
|
|
2UVD
 
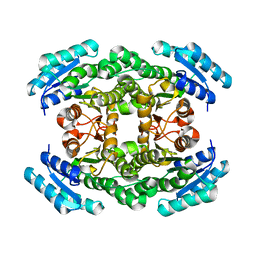 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2V25
 
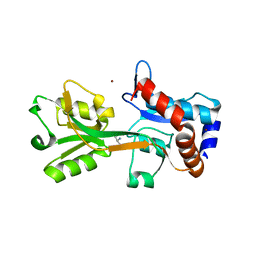 | | Structure of the Campylobacter jejuni antigen Peb1A, an aspartate and glutamate receptor with bound aspartate | | Descriptor: | ASPARTIC ACID, MAJOR CELL-BINDING FACTOR, ZINC ION | | Authors: | Muller, A, Dodson, E, del Rocio Leon-Kempis, M, Kelly, D.J, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Bacterial Virulence Factor with a Dual Role as an Adhesin and a Solute Binding-Protein: The Crystal Structure at 1.5 A Resolution of the Peb1A Protein from the Food-Borne Human Pathogen Campylobacter Jejuni
J.Mol.Biol., 372, 2007
|
|
2UVK
 
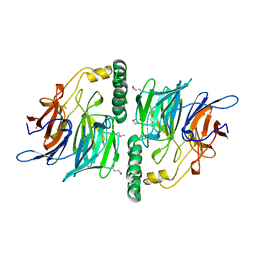 | | Structure of YjhT | | Descriptor: | YJHT | | Authors: | Muller, A, Severi, E, Wilson, K.S, Thomas, G.H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sialic Acid Mutarotation is Catalyzed by the Escherichia Coli Beta-Propeller Protein Yjht.
J.Biol.Chem., 283, 2008
|
|
2WAN
 
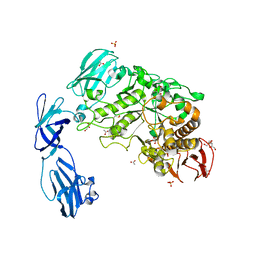 | | Pullulanase from Bacillus acidopullulyticus | | Descriptor: | ACETATE ION, GLYCEROL, PULLULANASE, ... | | Authors: | Turkenburg, J.P, Brzozowski, A.M, Svendsen, A, Borchert, T.V, Davies, G.J, Wilson, K.S. | | Deposit date: | 2009-02-10 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a Pullulanase from Bacillus Acidopullulyticus.
Proteins, 76, 2009
|
|
6ZMV
 
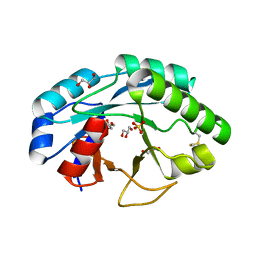 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
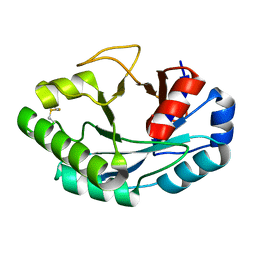 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
2C40
 
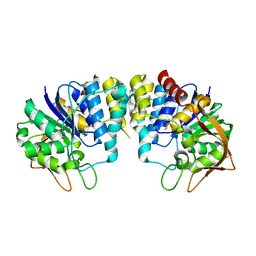 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | Descriptor: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | Authors: | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2C0S
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
2BTU
 
 | | Crystal structure of Phosphoribosylformylglycinamidine cyclo-ligase from Bacillus Anthracis at 2.3A resolution. | | Descriptor: | PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE | | Authors: | Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Lebedev, A.A, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-06-07 | | Release date: | 2006-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Phosphoribosylformylglycinamidine Cyclo-Ligase from Bacillus Anthracis at 2.3A Resolution.
To be Published
|
|
2C20
 
 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
2BZB
 
 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2C8J
 
 | | CRYSTAL STRUCTURE OF ferrochelatase HemH-1 from Bacillus anthracis, str. Ames | | Descriptor: | FERROCHELATASE 1 | | Authors: | Muller, A, Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-12-05 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ferrochelatase Hemh-1 from Bacillus Anthracis, Str. Ames
To be Published
|
|
2CHU
 
 | | CeuE in complex with mecam | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N',N''-[BENZENE-1,3,5-TRIYLTRIS(METHYLENE)]TRIS(2,3-DIHYDROXYBENZAMIDE), ... | | Authors: | Muller, A, Wilkinson, A.J, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2006-03-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An [{Fe(Mecam)}(2)](6-) Bridge in the Crystal Structure of a Ferric Enterobactin Binding Protein.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2CIC
 
 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
