4PPL
 
 | | Crystal structure of eCGP123 H193Q variant at pH 7.5 | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
4PPJ
 
 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
1MMI
 
 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4HA8
 
 | | Structure of Staphylococcus aureus biotin protein ligase in complex with biotin acetylene | | Descriptor: | (3aS,4S,6aR)-4-(hex-5-yn-1-yl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Yap, M, Wilce, M, Polyak, S, Soares da Costa, T, Tieu, W. | | Deposit date: | 2012-09-25 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Staphylococcus aureus biotin protein ligase in complex with biotin acetylene
To be Published
|
|
2IY9
 
 | | Crystal structure of the A-subunit of the AB5 toxin from E. coli | | Descriptor: | SUBA | | Authors: | Paton, A.W, Beddoe, T, Thorpe, C.M, Whisstock, J.C, Wilce, M.C.J, Rossjohn, J, Talbot, U.M, Paton, J.C. | | Deposit date: | 2006-07-13 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ab5 Subtilase Cytotoxin Inactivates the Endoplasmic Reticulum Chaperone Bip
Nature, 443, 2006
|
|
7SO0
 
 | | Crystal Structure of the Engineered Tick Evasin EVA-P974(F31A) Complexed to Human Chemokine CCL2 | | Descriptor: | C-C motif chemokine 2, Evasin P974 | | Authors: | Bhusal, R.P, Devkota, S.R, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8FJ2
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(C8) Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
7SCU
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of Tick Evasin EVA-AAM1001 complexed to human chemokine CCL7
To Be Published
|
|
8FK9
 
 | | Crystal Structure of the Tick Evasin EVA-ACA1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P991 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine recognition by the class A3 tick evasin EVA-ACA1001.
Protein Sci., 33, 2024
|
|
8FJ0
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(Y44A) Complexed to Human Chemokine CCL2 | | Descriptor: | C-C motif chemokine 2, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
1J0R
 
 | | Crystal structure of the replication termination protein mutant C110S | | Descriptor: | replication termination protein | | Authors: | Vivian, J.P, Hastings, A.F, Duggin, I.G, Wake, R.G, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2002-11-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The impact of single cysteine residue mutations on the replication terminator protein
Biochem.Biophys.Res.Commun., 310, 2003
|
|
8FK8
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7 | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
8FJ3
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL7(Y13A) | | Descriptor: | C-C motif chemokine 7, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
2NO4
 
 | | Crystal Structure analysis of a Dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2ZNM
 
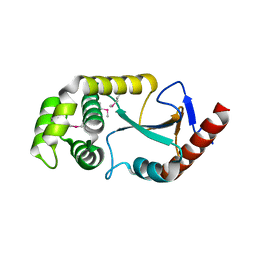 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
2NO5
 
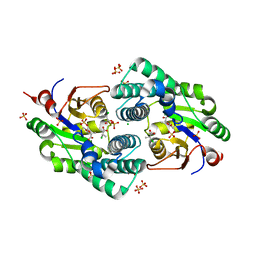 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | Descriptor: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | Authors: | Schmidberger, J.W, Wilce, M.C.J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
3VIC
 
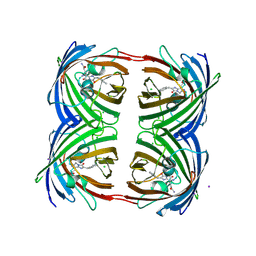 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
3VK1
 
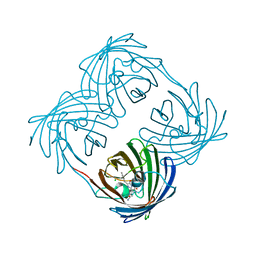 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
1BE7
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
1T3W
 
 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3BJX
 
 | |
3A3T
 
 | | The oxidoreductase NmDsbA1 from N. meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Bushell, S, Beddoe, T, Wilce, M.C.J, Byres, E, Boyle, T.P, Rimmer, K, Doak, B, Simpson, J.S, Graham, B, Heras, B, Kahler, C.M, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis
J.Mol.Biol., 394, 2009
|
|
1R0F
 
 | | Gallium-substituted rubredoxin | | Descriptor: | GALLIUM (III) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0H
 
 | | cobalt-substituted rubredoxin | | Descriptor: | COBALT (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
