7AYB
 
 | | Crystal Structure of wild type human mitochondrial 2-Enoyl Thioester Reductase (MECR) | | Descriptor: | ACETIC ACID, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, ... | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
3GZE
 
 | |
2CB8
 
 | | High resolution crystal structure of liganded human L-ACBP | | Descriptor: | 2-METHOXYETHANOL, ACYL-COA-BINDING PROTEIN, SULFATE ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-03 | | Release date: | 2006-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Crystal Structures of Unliganded and Liganded Human Liver Acbp Reveal a New Mode of Binding for the Acyl-Coa Ligand.
Proteins: Struct., Funct., Bioinf., 66, 2007
|
|
2FJ9
 
 | | High resolution crystal structure of the unliganded human ACBP | | Descriptor: | Acyl-CoA-binding protein, CHLORIDE ION, LEAD (II) ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of unliganded and liganded human liver ACBP reveal a new mode of binding for the acyl-CoA ligand.
Proteins, 66, 2006
|
|
1GUF
 
 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1, MITOCHONDRIAL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-25 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1GU7
 
 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1,MITOCHONDRIAL, GLYCEROL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
4C2K
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C2J
 
 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE, MITOCHONDRIAL, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BI9
 
 | | Crystal structure of wild-type SCP2 thiolase from Trypanosoma brucei. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
5AB7
 
 | |
5AB5
 
 | |
5AB6
 
 | |
5AB4
 
 | |
4BIA
 
 | | Crystal structure of SCP2 thiolase from Trypanosoma brucei: The C337A mutant. | | Descriptor: | 3-KETOACYL-COA THIOLASE, PUTATIVE | | Authors: | Harijan, R.K, Kiema, T.-R, Weiss, M.S, Michels, P.A.M, Wierenga, R.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Scp2-Thiolases of Trypanosomatidae, Human Pathogens Causing Widespread Tropical Diseases: The Importance for Catalysis of the Cysteine of the Unique Hdcf Loop.
Biochem.J., 455, 2013
|
|
4U1A
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase helix-10 deletion mutant (ISOB-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
4U18
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase (ISO-ECI2) | | Descriptor: | CHLORIDE ION, Enoyl-CoA delta isomerase 2, mitochondrial, ... | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
4U19
 
 | | Crystal structure of human peroxisomal delta3,delta2, enoyl-CoA isomerase V349A mutant (ISOA-ECI2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA delta isomerase 2 | | Authors: | Onwukwe, G.U, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Human Delta (3) , Delta (2) -enoyl-CoA isomerase, type 2: a structural enzymology study on the catalytic role of its ACBP domain and helix-10.
Febs J., 282, 2015
|
|
6TNM
 
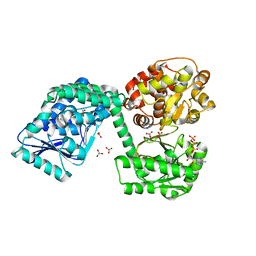 | | E. coli aerobic trifunctional enzyme subunit-alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Fatty acid oxidation complex subunit alpha, GLYCEROL, ... | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2019-12-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex.
J.Struct.Biol., 210, 2020
|
|
6EVO
 
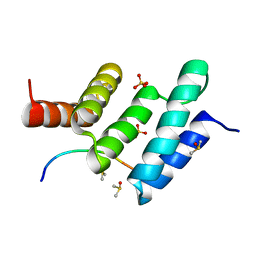 | | Crystal structure the peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complexed with Pro-Pro-Gly-Pro-Arg-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ARG-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
3ZWA
 
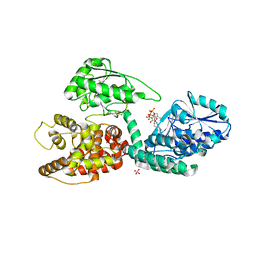 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-HEXANOYL-COA | | Descriptor: | (S)-3-Hydroxyhexanoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
6EVM
 
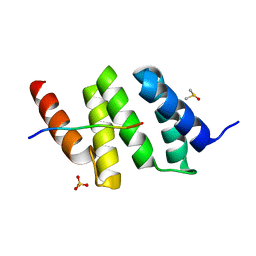 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
6EVN
 
 | | Crystal structure of peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase complex with Pro-Pro-Gly-Pro-Ala-Gly-Pro-Pro-Gly. | | Descriptor: | DIMETHYL SULFOXIDE, PRO-PRO-GLY-PRO-ALA-GLY-PRO-PRO-GLY, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
3ZW8
 
 | | Crystal Structure Of Rat Peroxisomal Multifunctional Enzyme Type 1 (rpMFE1) In Apo Form | | Descriptor: | GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, SULFATE ION | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWC
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 3S-HYDROXY-DECANOYL-COA | | Descriptor: | (S)-3-HYDROXYDECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
3ZWB
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE 1 (RPMFE1) COMPLEXED WITH 2TRANS-HEXENOYL-COA | | Descriptor: | (2E)-Hexenoyl-CoA, GLYCEROL, PEROXISOMAL BIFUNCTIONAL ENZYME, ... | | Authors: | Kasaragod, P, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Isomerase and Hydratase Reaction Mechanism of the Crotonase Active Site of the Multifunctional Enzyme (Type-1), as Deduced from Structures of Complexes with 3S-Hydroxy- Acyl-Coa.
FEBS J., 280, 2013
|
|
