2GCE
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-IBUPROFENOYL-COENZYME A, (S)-IBUPROFENOYL-COENZYME A, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2J24
 
 | | The functional role of the conserved active site proline of triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Casteleijn, M.G, Alahuhta, M, Groebel, K, El-Sayed, I, Augustyns, K, Lambeir, A.M, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2006-08-16 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional Role of the Conserved Active Site Proline of Triosephosphate Isomerase.
Biochemistry, 45, 2006
|
|
1M1O
 
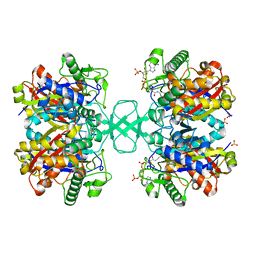 | | Crystal structure of biosynthetic thiolase, C89A mutant, complexed with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
2GCI
 
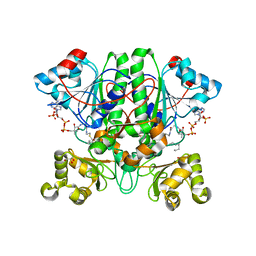 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2JIG
 
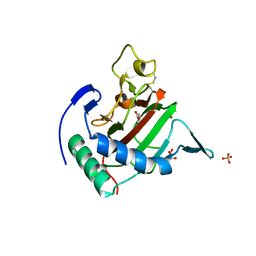 | | Crystal structure of Chlamydomonas reinhardtii prolyl-4 hydroxylase type I complexed with zinc and pyridine-2,4-dicarboxylate | | Descriptor: | GLYCEROL, PROLYL-4 HYDROXYLASE, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
1NL7
 
 | |
1M4T
 
 | | Biosynthetic thiolase, Cys89 butyrylated | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
2JIJ
 
 | | Crystal structure of the apo form of Chlamydomonas reinhardtii prolyl- 4 hydroxylase type I | | Descriptor: | CHLORIDE ION, PROLYL-4 HYDROXYLASE | | Authors: | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
1GYR
 
 | | Mutant form of enoyl thioester reductase from Candida tropicalis | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, SULFATE ION | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-04-29 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
1SHF
 
 | |
1DCI
 
 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
1E15
 
 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1PJH
 
 | |
2V5L
 
 | | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: as Observed in a New Crystal Form: Implications for the Reaction Mechanism | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E.M, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: As Observed in a New Crystal Form: Implications for the Reaction Mechanism
Proteins: Struct.,Funct., Genet., 16, 1993
|
|
1PXT
 
 | |
1SSG
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW0
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SW3
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant T175V | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1N55
 
 | |
1SSD
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1SPQ
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
2J27
 
 | | The functional role of the conserved active site proline of triosephosphate isomerase. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Casteleijn, M.G, Alahuhta, M, Groebel, K, El-Sayed, I, Augustyns, K, Lambeir, A.M, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2006-08-16 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Functional Role of the Conserved Active Site Proline of Triosephosphate Isomerase.
Biochemistry, 45, 2006
|
|
