2QV7
 
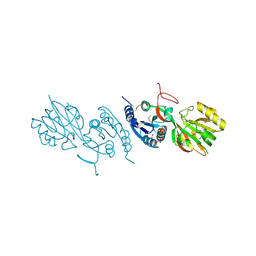 | | Crystal Structure of Diacylglycerol Kinase DgkB in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Diacylglycerol Kinase DgkB, MAGNESIUM ION | | Authors: | Miller, D.J, Jerga, A, Rock, C.O, White, S.W. | | Deposit date: | 2007-08-07 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the Staphylococcus aureus DgkB Structure Reveals a Common Catalytic Mechanism for the Soluble Diacylglycerol Kinases.
Structure, 16, 2008
|
|
2QVL
 
 | |
1K23
 
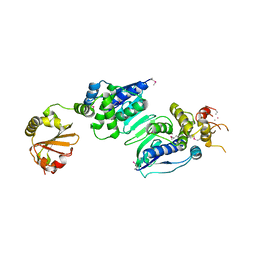 | | Inorganic Pyrophosphatase (Family II) from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase | | Authors: | Ahn, S, Milner, A.J, Futterer, K, Konopka, M, Ilias, M, Young, T.W, White, S.A. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The "open" and "closed" structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii.
J.Mol.Biol., 313, 2001
|
|
1K20
 
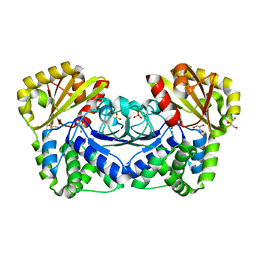 | | Inorganic Pyrophosphatase (family II) from Streptococcus gordonii at 1.5 A resolution | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Manganese-dependent inorganic pyrophosphatase, ... | | Authors: | Ahn, S, Milner, A.J, Futterer, K, Konopka, M, Ilias, M, Young, T.W, White, S.A. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The "open" and "closed" structures of the type-C inorganic pyrophosphatases from Bacillus subtilis and Streptococcus gordonii.
J.Mol.Biol., 313, 2001
|
|
1A32
 
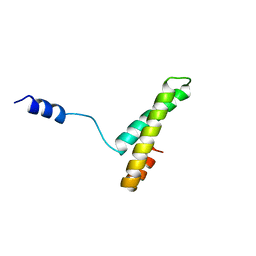 | |
1LRW
 
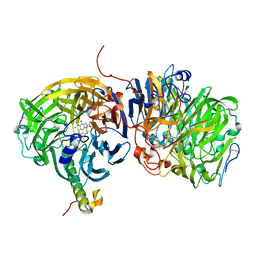 | | Crystal structure of methanol dehydrogenase from P. denitrificans | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, methanol dehydrogenase subunit 1, ... | | Authors: | Xia, Z.-X, Dai, W.-W, He, Y.-N, White, S.A, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-05-16 | | Release date: | 2003-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of methanol dehydrogenase from Paracoccus denitrificans and molecular modeling of its interactions with cytochrome c-551i
J.Biol.Inorg.Chem., 8, 2003
|
|
1MMS
 
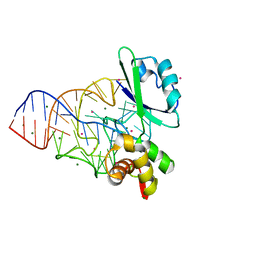 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
1CVX
 
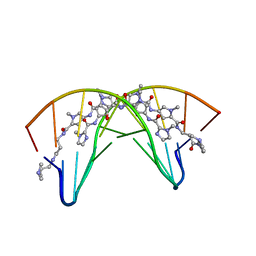 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1CVY
 
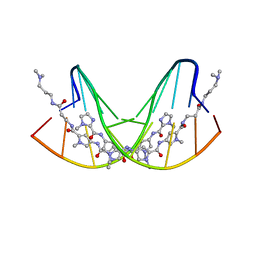 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYPYPYBETADP)2 BOUND TO CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
1KAF
 
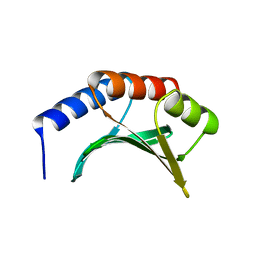 | | DNA Binding Domain Of The Phage T4 Transcription Factor MotA (AA105-211) | | Descriptor: | Transcription regulatory protein MOTA | | Authors: | Li, N, Sickmier, E.A, Zhang, R, Joachimiak, A, White, S.W. | | Deposit date: | 2001-11-01 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif.
Mol.Microbiol., 43, 2002
|
|
1ICV
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1ICU
 
 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
1ICR
 
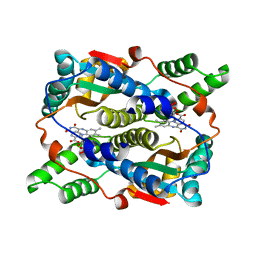 | | THE STRUCTURE OF ESCHERICHIA COLI NITROREDUCTASE COMPLEXED WITH NICOTINIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Lovering, A.L, Hyde, E.I, Searle, P.F, White, S.A. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 A, 1.8 A and 2.4 A resolution.
J.Mol.Biol., 309, 2001
|
|
7KNR
 
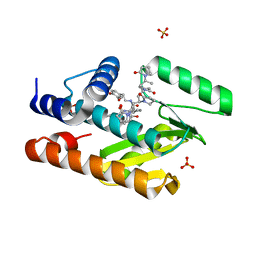 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988558
To Be Published
|
|
7KNY
 
 | | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, White, S.W. | | Deposit date: | 2020-11-06 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The crystal structure of the I38T mutant PA endonuclease (2009/H1N1/California) in complex with SJ000988528
To Be Published
|
|
1I01
 
 | |
1I1S
 
 | | SOLUTION STRUCTURE OF THE TRANSCRIPTIONAL ACTIVATION DOMAIN OF THE BACTERIOPHAGE T4 PROTEIN MOTA | | Descriptor: | MOTA | | Authors: | Li, N, Zhang, W, White, S.W, Kriwacki, R.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA.
Biochemistry, 40, 2001
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
7KOP
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539 | | Descriptor: | 2-(2-fluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Seetharaman, J, White, S.W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with SJ000988539
To Be Published
|
|
7K87
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease in complex with SJ000986436 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7KL3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D bound to RNA oligomer AG*CAUC (*uncleaveable bond, -UC disordered) | | Descriptor: | 5'-O-sulfocytidine, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cuypers, M.G, Kumar, G, White, S.W. | | Deposit date: | 2020-10-28 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7K0W
 
 | |
7KBC
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, GLYCEROL, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-10-02 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7KAF
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-09-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
