6O5M
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 10bb | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design, Synthesis, and Biological Evaluation of (2-(1H-Indol-3-yl)-1H-imidazol-4-yl)(3,4,5-trimethoxyphenyl) Methanone (ABI-231) Analogues Targeting the Colchicine Binding Site in Tubulin.
J.Med.Chem., 62, 2019
|
|
3H24
 
 | |
3SQ5
 
 | |
5DES
 
 | | 2009 H1N1 PA endonuclease domain | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YWO
 
 | | Mercuric reductase from Metallosphaera sedula | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase | | Authors: | Artz, J.H, Zadvornyy, O.A, White, S, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Biochemical and Structural Properties of a Thermostable Mercuric Ion Reductase from Metallosphaera sedula.
Front Bioeng Biotechnol, 3, 2015
|
|
3H23
 
 | |
3H2E
 
 | | Structural Studies of Pterin-Based Inhibitors of Dihydropteroate Synthase | | Descriptor: | 1,3-dimethyl-2,4,7-trioxo-1,2,3,4,7,8-hexahydropteridine-6-carbaldehyde, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of pterin-based inhibitors of dihydropteroate synthase.
J.Med.Chem., 53, 2010
|
|
3H21
 
 | | Structural Studies of Pterin-Based Inhibitors of Dihydropteroate Synthase | | Descriptor: | (2R)-2-(7-amino-1-methyl-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)propanoic acid, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural studies of pterin-based inhibitors of dihydropteroate synthase.
J.Med.Chem., 53, 2010
|
|
3H2C
 
 | |
3H2N
 
 | | Structural Studies of Pterin-Based Inhibitors of Dihydropteroate Synthase | | Descriptor: | (6R)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, Dihydropteroate synthase, SULFATE ION | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of pterin-based inhibitors of dihydropteroate synthase.
J.Med.Chem., 53, 2010
|
|
3H2A
 
 | |
3H2M
 
 | |
5JLT
 
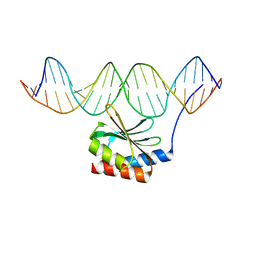 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
1YKI
 
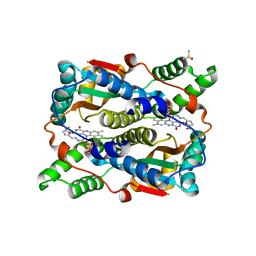 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | Descriptor: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
2F9T
 
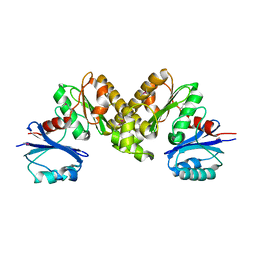 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | Pantothenate Kinase | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
2F9W
 
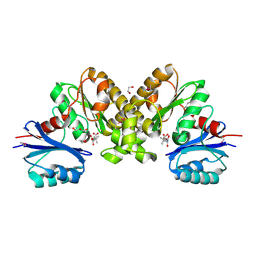 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
5EGA
 
 | | 2009 H1N1 PA endonuclease domain in complex with an N-acylhydrazone inhibitor | | Descriptor: | 3,4,5-tris(oxidanyl)-~{N}-[(~{E})-[3,4,5-tris(oxidanyl)phenyl]methylideneamino]benzamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-10-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-acylhydrazone inhibitors of influenza virus PA endonuclease with versatile metal binding modes.
Sci Rep, 6, 2016
|
|
6BR1
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
1PT9
 
 | | Crystal Structure Analysis of the DIII Component of Transhydrogenase with a Thio-Nicotinamide Nucleotide Analogue | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
6BS2
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRF
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRY
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
1PKP
 
 | |
1WHI
 
 | | RIBOSOMAL PROTEIN L14 | | Descriptor: | RIBOSOMAL PROTEIN L14 | | Authors: | Davies, C, White, S.W, Ramakrishnan, V. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of ribosomal protein L14 reveals an important organizational component of the translational apparatus.
Structure, 4, 1996
|
|
4JUL
 
 | | Crystal structure of H5N1 influenza virus hemagglutinin, clade 2.3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Coop, T, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2013-03-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7927 Å) | | Cite: | To be published
To be Published
|
|
